This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Breakthrough in base editing enhances genetic engineering, trait evolution in watermelon
Cytosine and adenosine base editors (CBEs and ABEs) are integral to molecular breeding, enable precise modification of single-nucleotide polymorphisms (SNPs) in plants, which are crucial for agronomic traits and species evolution. Despite their success in crops like rice, maize, and watermelon, their scope is limited to these two types of base substitutions.
Recent developments in mammalian cells with CGBE (C-to-G base editor) show promise for expanding this capacity. However, challenges persist in achieving high efficiency and broader application in plant species, particularly in crops like watermelon where SNP substitutions significantly impact trait evolution. The current research focus is on developing efficient CGBE tailored for watermelon, aiming to broaden the scope of base editing in plant molecular breeding.
The article, titled "Developing a highly efficient CGBE base editor in watermelon," was published in Horticulture Research.
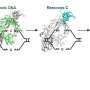
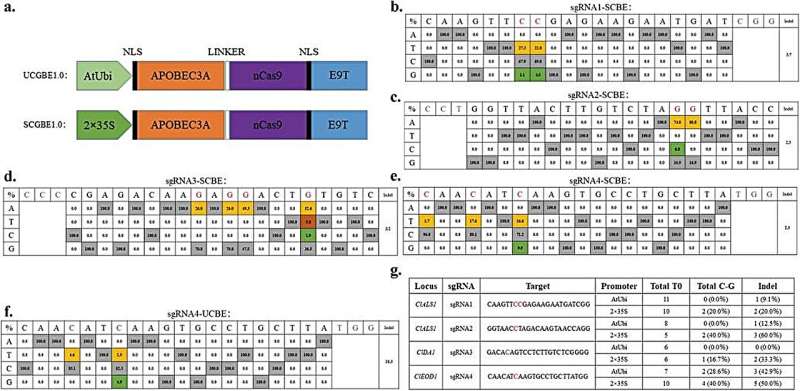
In this study, a two-step optimization process was employed to develop a Cytosine to Guanine Base Editor (CGBE) specifically tailored for watermelon, integrating human APOBEC3A (hA3A) deaminase and Arabidopsis UNG. Initially, hA3A-CBEs were designed to induce C-to-G base substitutions along with C-to-T editing. Various promoters, such as AtUbi, 2×35S, and AtRps5a, were tested to drive Arabidopsis codon-optimized hA3A-nCas9 (D10A)-UGI fusion protein expression.
Transgenic watermelon plants were generated using four sgRNAs targeting three watermelon genes (ClALS1, ClDA1, and ClEOD1). The results showed that the AtUbi and 2×35S promoter-based CBEs achieved high C-to-T editing efficiencies (27.0% and 93.4%, respectively), while some samples also exhibited C-to-G transversions, likely due to the recognition and excision of C-to-U mutations by endogenous UNG.
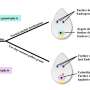
To improve the applicability of base editing techniques in watermelon, the study developed an efficient CGBE editor (SCGBE2.0) by removing the uracil glycosylase inhibitor (UGI) unit from the commonly used hA3A-CBE and adding a uracil-DNA glycosylase (UNG) component.
Results obtained from stably transformed watermelon plants showed that SCGBE2.0 was effective in inducing C-G mutations at the C5-C9 position in 43.2% of transgenic plants (maximum base conversion efficiency of 46.1%) and C-A mutations at the C4 position in 23.5% of transgenic plants (maximum base conversion efficiency of 45.9%).
This editor preferred NGG and NAG PAM sequences, expanding the possibilities for site-preferred C-to-G/A conversions in watermelon. In addition, future optimizations of SCGBE2.0 should focus on narrowing the editing window and reducing indel formation to enhance the precision and efficiency of watermelon genome editing.
Overall, this novel editing system achieved efficient C-to-G/A mutations in watermelon, providing an efficient base editing tool for precise base modification and site-directed saturation mutagenesis in watermelon. This breakthrough in CGBE development not only enhances the scope of molecular breeding in watermelon but also offers new avenues for research in other dicotyledonous plants.
More information: Dong Wang et al, Developing a highly efficient CGBE base editor in watermelon, Horticulture Research (2023). DOI: 10.1093/hr/uhad155
Journal information: Horticulture Research
Provided by TranSpread