This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New approach to predicting binding properties of PFAS chemicals and human PPARα
Per- and polyfluoroalkyl substances (PFAS) are widely used in various products as water-repellents and stain-resistant coatings. PFAS are called "forever chemicals" due to their exceptional thermal and chemical stability, and have been found globally in the environment, humans, and wildlife.
Long-chain perfluoroalkyl acids, including perfluorooctanoic acid (PFOA) and perfluorooctanesulfonic acid (PFOS), are persistent, bioaccumulative, and toxic. Globally, PFOA- and PFOS-related substances are regulated by the Stockholm Convention on Persistent Organic Pollutants (POPs).
A key toxicological aspect of PFAS, especially PFOA and PFOS, is their disruption of lipid metabolism through interaction with the PPARα, essential in lipid metabolism, energy balance, and cell differentiation. PFAS binding to PPARα disrupts signaling pathways, causing various biological effects. However, the potential hazards (e.g., bioactivity, bioaccumulation, and toxicity) of thousands of PFAS types, including next-generation alternative PFAS is limited.
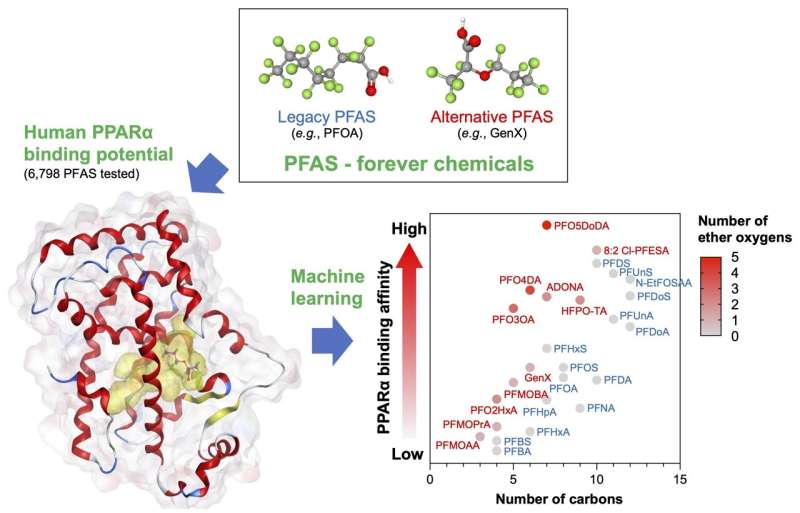
In a study published in Environmental Science & Technology, researchers developed an explainable machine learning approach to predict the binding affinity of PFAS-PPARα.
They obtained SMILES data for 6,798 PFAS from the U.S. EPA database and used Molecular Operating Environment (MOE) to calculate 206 molecular descriptors and binding affinity (i.e., S-score) to PPARα for each PFAS. Results revealed that 4,089 PFAS exhibited S-scores lower than those of both PFOA (S-score = -5.03 kcal/mol) and PFOS (S-score = -5.09 kcal/mol).
Through the systematic and objective selection of important molecular descriptors, the team developed a machine learning model with good predictive performance using only three descriptors (R2=0.72). The molecular size (b_single) and electrostatic properties (BCUT_PEOE_3 and PEOE_VSA_PPOS) are important for PPARα-PFAS binding. Alternative PFAS are considered safer than their legacy predecessors.
However, the researchers found that alternative PFAS with many carbon atoms and ether groups exhibited a higher binding affinity for PPARα than legacy PFOA and PFOS. The novel approach outperforms traditional QSAR and machine learning approaches in terms of interpretability, thereby providing deeper insight into the molecular mechanism of PFAS toxicity.
In the present study, the machine learning model successfully predicted the binding affinity of PFAS to human PPARα and predicted key molecular characteristics in the binding. Although this study focused on PFAS-PPARα binding and was limited to ligand-receptor binding, the team's approach is also relevant to other ligand-receptor binding and other structure-property relationship studies.
Future research could improve the accuracy of toxicity predictions by incorporating more features. Such studies would involve not only the structural details of PFAS but also information about downstream signal transduction pathways, thereby potentially enabling more precise toxicity predictions. However, limitations exist.
The researchers focused on the interaction with PPARα, whereas PFAS could induce toxicity through other receptors. Notably, a high binding score does not always reflect toxicity. Thus, the actual toxicity must be experimentally verified. Despite these limitations, their method allows for rapid, cost-effective PFAS screening, providing a preliminary understanding their potential toxicity and guiding further in-depth experimental investigations.
More information: Kazuhiro Maeda et al, Elucidating Key Characteristics of PFAS Binding to Human Peroxisome Proliferator-Activated Receptor Alpha: An Explainable Machine Learning Approach, Environmental Science & Technology (2023). DOI: 10.1021/acs.est.3c06561
Journal information: Environmental Science & Technology
Provided by Ehime University