This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Textbook knowledge turned on its head: 3-in-1 microorganism discovered
A team of researchers has now been able to show that there is an incredibly high biodiversity of environmentally relevant microorganisms in nature. This diversity is at least 4.5 times greater than previously known. The researchers recently published their findings in the journals Nature Communications and FEMS Microbiology Reviews.
The hidden world of microorganisms is often overlooked, even though many climate-relevant processes are influenced by microorganisms, often associated with an incredible diversity of species within the groups of bacteria and archaea ("primitive bacteria").
For example, sulfate-reducing microorganisms convert a third of the organic carbon in marine sediments into carbon dioxide. This produces toxic hydrogen sulfide. On the positive side, sulfur-oxidizing microorganisms quickly use this as an energy source and render it harmless.
"These processes also play an important role in lakes, bogs and even in the human gut to keep nature and health in balance," says Prof. Michael Pester, Head of the Department of Microorganisms at the Leibniz Institute DSMZ and Professor at the Institute of Microbiology at Technische Universität Braunschweig. A study examined the metabolism of one of these novel microorganisms in more detail, revealing a multifunctionality that was previously unattainable.
Microorganisms stabilize ecosystems
The sulfur cycle is one of the most important and oldest biogeochemical cycles on our planet. At the same time, it is closely linked to the carbon and nitrogen cycles, underlining its importance. It is mainly driven by sulfate-reducing and sulfur-oxidizing microorganisms. On a global scale, sulfate reducers convert about a third of the organic carbon that reaches the seafloor each year. In return, sulfur oxidizers consume about a quarter of the oxygen in marine sediments.
When these ecosystems become unbalanced, the activities of these microorganisms can rapidly lead to oxygen depletion and the accumulation of toxic hydrogen sulfide. This leads to the formation of 'dead zones' where animals and plants can no longer survive. This not only causes economic damage, for example to fisheries, but also social damage through the destruction of important local recreational areas. It is therefore important to understand which microorganisms keep the sulfur cycle in balance and how they do this.
The published results show that the species diversity of sulfate-reducing microorganisms includes at least 27 phyla (strains). Previously, only six phyla were known. By comparison, 40 phyla are currently known in the animal kingdom, with vertebrates belonging to only one phylum, the Chordata.
Newly discovered multifunctional bacterial species
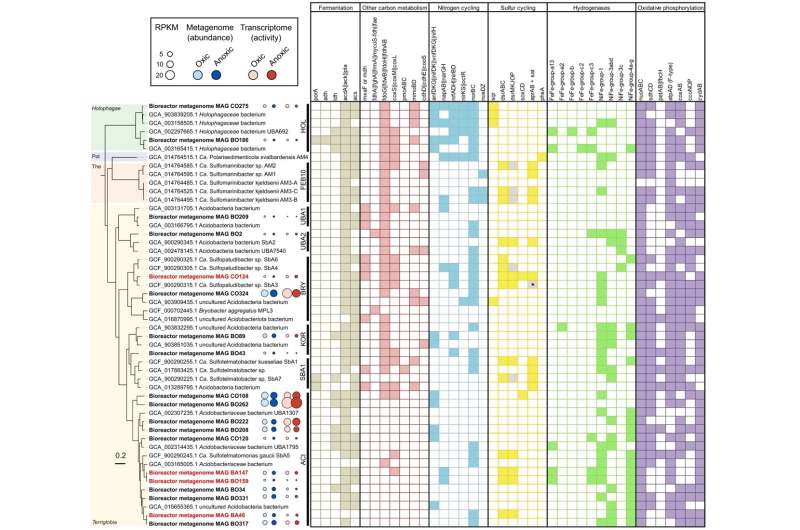
The researchers were able to assign one of these novel "sulfate reducers" to the little researched phylum of acidobacteriota and to study it in a bioreactor.
Using cutting-edge methods from environmental microbiology, they were able to show that these bacteria can obtain energy from both sulfate reduction and oxygen respiration. These two pathways are normally mutually exclusive in all known microorganisms. At the same time, the researchers were able to show that the sulfate-reducing acidobacteriota can break down complex plant carbohydrates such as pectin—another previously unknown property of "sulfate reducers."
The researchers have thus turned textbook knowledge on its head. They show that complex plant compounds can be degraded under oxygen exclusion not only by the coordinated interaction of different microorganisms, as previously thought, but also by a single bacterial species via a shortcut.
Another new finding is that these bacteria can use both sulfate and oxygen for this purpose. Researchers at the DSMZ and Technische Universität Braunschweig are currently investigating how the new findings affect the interplay of the carbon and sulfur cycles and how they are linked to climate-relevant processes.
More information: Stefan Dyksma et al, Oxygen respiration and polysaccharide degradation by a sulfate-reducing acidobacterium, Nature Communications (2023). DOI: 10.1038/s41467-023-42074-z
Muhe Diao et al, Global diversity and inferred ecophysiology of microorganisms with the potential for dissimilatory sulfate/sulfite reduction, FEMS Microbiology Reviews (2023). DOI: 10.1093/femsre/fuad058
Journal information: Nature Communications
Provided by Technical University of Braunschweig