This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Researchers supply significant genomic insight into tar spot on corn
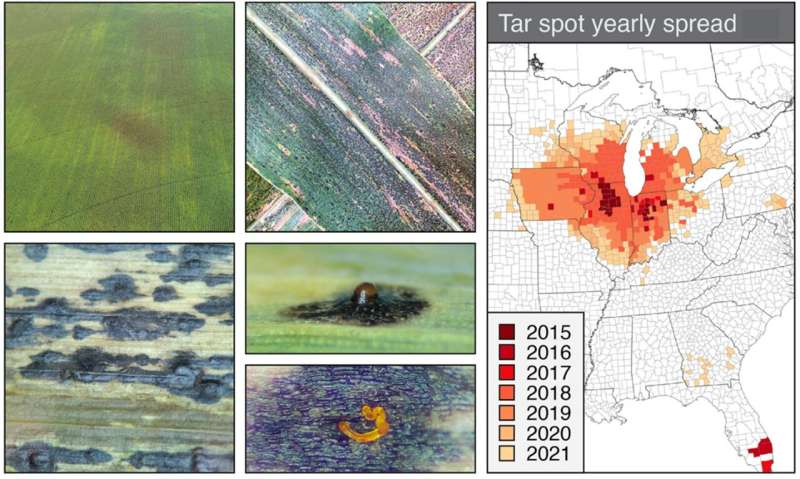
First reported in 2015, tar spot is an emerging disease on corn that has rapidly spread across the United States and Canada, causing tremendous yield loss estimated at $1.2 billion in 2021 alone. Tar spot gets its name from its iconic symptoms that resemble the splatter of "tar" on corn leaves, but these spots are in fact brown lesions formed by the fungal pathogen Phyllachora maydis.
This destructive pathogen is challenging to research because it cannot survive outside its plant host; therefore, little information is currently known about the mechanisms that contribute to its disease cycle including spore formation, reproduction, and plant infection.
Researchers have consequently come to rely on genome sequencing to identify what genes are present and predict what roles they play during disease development. However, the current draft genome is largely incomplete, and although genes are present in the fungus, they may not be expressed during infection.
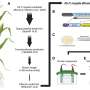
To address this, a recent study led by Joshua MacCready and Emily Roggenkamp from Martin I. Chilvers's lab at Michigan State University generated a high-quality genome for P. maydis and provided the first global gene expression analysis of tar spot disease infection. The study, published in the journal Molecular Plant-Microbe Interactions, found that the fungus undergoes sexual reproduction early on in lesion formation, and this requires two individuals of different mating types.
This finding is significant because sexual reproduction allows the fungus more genetic flexibility to evolve, resulting in potentially more virulent or fungicide resistant isolates. Like many fungal pathogens, P. maydis relies on the secretion of proteins, called effectors, to manipulate and overcome plant immunity.
This study identified over 100 effectors, many of which were found to be unique to this fungal pathogen and highly expressed during infection, suggesting that these proteins play a novel role during infection and warrant further investigation to determine their function.
The genome and gene expression analysis provides a valuable resource for other researchers studying tar spot on corn. Chilvers states, "This study lays the groundwork for future studies to examine effectors, pathogenesis, and the lifecycle of this pathogen." Armed with an improved understanding about this fungus, these researchers aim to implement improved disease management tactics for this impactful pathogen.
More information: Joshua S. MacCready et al, Elucidating the Obligate Nature and Biological Capacity of an Invasive Fungal Corn Pathogen, Molecular Plant-Microbe Interactions (2023). DOI: 10.1094/MPMI-10-22-0213-R
Journal information: Molecular Plant-Microbe Interactions
Provided by American Phytopathological Society