This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
How green algae count cell divisions illuminates key step needed for the evolution of multicellular life
An international research team led by James Umen, Ph.D., member, Donald Danforth Plant Science Center has made an unexpected discovery of a biased counting mechanism used by the single-celled green alga Chlamydomonas to control cell division. Chlamydomonas cells can grow very large and then divide multiple times in succession.
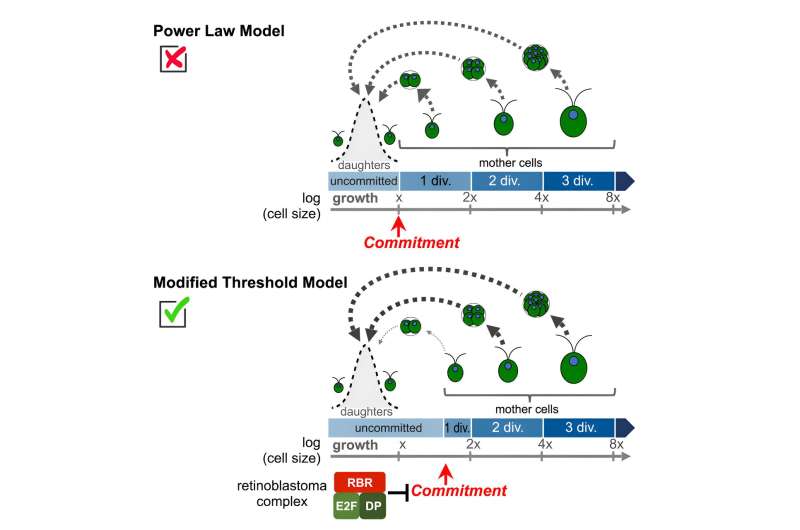
The team found that the number of divisions a mother cell undertakes to restore its daughters to the correct starting size deviates from the mathematical optimum that was assumed to dictate this process. Instead, mother cells almost never divided just one time—they either didn't divide at all or divided two or more times.
This unexpected bias against a single division has important implications for understanding the evolution of multicellular life and provides a new avenue for engineering algal cells for improved yields of biofuel and high value products. The article describing these findings titled, "A Cell-Based Model for Size Control in the Multiple Fission Alga Chlamydomonas reinhardtii," was published in the journal Current Biology.
Chlamydomonas cells, like those of many other algae and single-celled protists, can grow very large before they divide. This atypical growth and division pattern lets them make optimal use of light and nutrients, but also creates a problem in size control: Under some conditions cells will just barely double their size before it is time to divide and only need to divide once; but under favorable conditions the same cell might grow more than ten times its starting size and would need to divide multiple times in succession to produce daughters of the correct size.
This size variability presents a conundrum which was solved by the evolution of a mechanism in Chlamydomonas that enables cells to assess their size and count out the correct number of cell divisions. "It was always assumed that the division pattern was dictated by a simple relationship between mother cell size and number of divisions, and models that assume this simple relationship can accurately predict the behaviors of entire cell populations," said Umen.
"But by looking at division behaviors of thousands of individual cells of varying sizes we found an unanticipated dearth of cells dividing just once." Instead, cells that should have divided once opted to not divide at all, and most cells only became able to divide when they had more than doubled in size.
Faced with this unexpected result, team members Abhyudai Singh, Ph.D., professor, University of Delaware and César Augusto Vargas-García, Ph.D., analytics team leader, AGROSAVIA -Corporación colombiana de investigación agropecuaria, Bogotá, Colombia, used mathematical modeling to come up with a more accurate predictive model for the behavior of the cells, while the research team at Danforth Center, spearheaded by Dianyi Liu, Ph.D., postdoctoral associate, dug deeper to understand the genetic mechanisms that produced the observed counting bias.
The team discovered that a well-known, but still poorly understood genetic mechanism for controlling cell division found in algae, plants and humans—called the retinoblastoma tumor suppressor pathway—plays a critical role in preventing just one division.
"While we are just at the start of understanding how the retinoblastoma pathway works in algae, the discovery of a mechanism for introducing bias in cell division behavior immediately suggests a way that cells modified their division behavior as an important step in the evolution of multicellular life," according to Liu.
Multicellular relatives of Chlamydomonas not only skip the option to undergo one division but can prevent division until they have grown many times in size. This enables a single cell to rapidly produce a whole new multicellular individual with hundreds or even thousands of cells, an ability that is critical for fitness and survival.
"The bias against one division we observed in Chlamydomonas was very likely present in direct ancestors of its multicellular relatives and was further amplified as they evolved greater size and complexity," speculates Umen. While it remains unclear why the cells evolved a bias against dividing just once, the knowledge of this mechanism and its genetic control has practical implications in algal biotechnology where cell size can impact yields of high value products and even susceptibility to predation of algae by filter feeders in open pond cultures.
Looking forward, the team is now focused on understanding and modeling the specific mechanisms used by the retinoblastoma pathway to alter cell division behavior in algae, work that may lead to advances in algal biotechnology and even shed light on how the retinoblastoma pathway keeps human cells from developing cancer and prevents plant cells from dividing at the wrong time and place.
More information: Dianyi Liu et al, A cell-based model for size control in the multiple fission alga Chlamydomonas reinhardtii, Current Biology (2023). DOI: 10.1016/j.cub.2023.10.023
Journal information: Current Biology
Provided by Donald Danforth Plant Science Center