This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
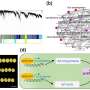
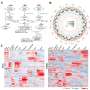
Comprehensive analysis of the telomere-to-telomere genome of soybean cultivar ZH13

Soybean (Glycine max) is one of the most crucial oil and protein crops, and contributes to more than a quarter of the protein utilized in both human food and animal feed. It is widely acknowledged that the cultivated soybean emerged through the domestication of its annual ancestor in the Yellow River basin. Therefore, the exploration of genetic resources within the origin region bears immense significance in advancing the global frontiers of soybean breeding.
In comparison to the cultivar of Williams 82, Zhonghuang 13 (ZH13) boasts higher genetic diversity and ecological type of origin reign. Furthermore, ZH13 is an ideal variety in the breeding strategy called "Potalaization," which allows breeding of novel widely adapted soybean varieties through the use of multiple molecular tools in existing elite widely adapted varieties. To date, however, soybean genome analyses are incomplete and contain many gaps, which have so far limited in-depth investigations into its properties.
To address this, a joint team of researchers from China, including senior author and co-corresponding author Dr. Yadong Wang from the Center for Bioinformatics, School of Computer Science and Technology at the Harbin Institute of Technology, and co-corresponding author Dr. Tianfu Han from the Institute of Crop Sciences at the Chinese Academy of Agricultural Sciences, conducted a telomere-to-telomere (T2T) assembly of the Chinese soybean cultivar ZH13, termed ZH13-T2T. The study was published in The Crop Journal.
"Imagine you have a giant jigsaw puzzle, but it's missing some pieces. This puzzle is like the genetic code, or the 'recipe,' of ZH13. The missing pieces are like gaps in our understanding of this recipe," explained by Dr. Yang Hu, co-corresponding author of the study. "In this effort, we used a super-advanced and precise method to find and fit in all those missing pieces. With this powerful tool, we could see everything—even the tricky parts that were hidden before."
The researchers used a multi-assembler approach to minimize biases and enhance assembly accuracy. Their assembly spanned 1,015,024,879 base pairs (bp), effectively filling in all the gaps from the previous analyses. In the process, they identified more than 50,000 protein-coding genes, of which 707 are novel. ZH13-T2T was found to have longer chromosomes, 421 not-aligned regions (NARs), 112 structure variations (SVs), and a significant expansion of repetitive elements compared to earlier analyses.
"We've delivered the first complete Chinese soybean cultivar T2T genome," said Dr. Bo Liu, the other co-corresponding author. The complete, accurate genome sequence of the ZH13 cultivar can now be used to identify crucial genes, and genetic variants linked to desirable traits."
This information would also contribute to accelerating soybean breeding programs to develop new cultivars with specific enhanced traits, crop yields, improved resistance to pests and diseases, and adaptability to different regions and climates.
"For instance, researchers can use the genomic information to manipulate specific genes that encode improvements in soybean traits, such as photothermal adaptability, oil content, protein quality, or tolerance to abiotic and biotic stressors," concluded by Baiquan Sun, an author of the study.
More information: Anqi Zhang et al, A telomere-to-telomere genome assembly of Zhonghuang 13, a widely-grown soybean variety from the original center of Glycine max, The Crop Journal (2023). DOI: 10.1016/j.cj.2023.10.003
Provided by KeAi Communications Co.