September 14, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Team develops technique for building DNA-based programmable gate arrays
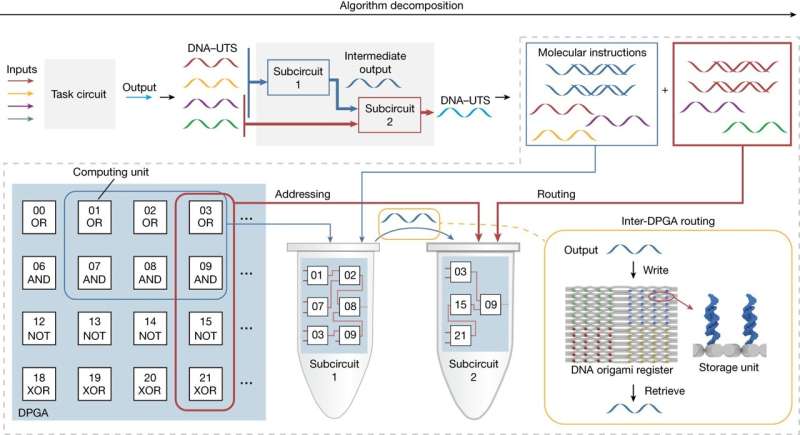
A team of chemists and chemical engineers at Shanghai Jiao Tong University, working with a colleague from the Chinese Academy of Sciences, both in China, has built a DNA-based programmable gate array for use in general-purpose DNA computing. In their study, reported in the journal Nature, the group overcame obstacles that had hindered the development of multipurpose DNA-based circuits and created circuits using their new process.
In 1994, Leonard Adleman won the Turing award for his proposed use of DNA base-pairing to create a biocomputing device. Since that time, many such devices have been created. But until now, any given device could do only one thing. In this new effort, the team in China has overcome problems faced by other researchers working on making such devices more universal by developing a technique for creating a field-programmable gate array using DNA, which they describe as a DNA-based programmable gate array (DPGA).
The work involved adding strands of DNA to a tube containing a buffer fluid. They then induced chemical reactions that forced the strands together in desired ways, creating longer strands that together made up a DPGA. The team then added fluorescence markers to allow for viewing and keeping track of circuit formation. Testing showed that a single DPGA could be designed in a way that allowed for the creation of 100 billion unique circuits simply by adding specific amounts of short molecules to the DNA strands.
In testing their DPGAs, the researchers found that by connecting just three of them together, they were able to build a circuit capable of solving quadratic equations or square roots. They note that numbers could be added for computing such formulas by adding molecules of a specific shape. Output from the DPGAs was done by studying the molecules that were produced by the final reaction using the fluorescent markers.
The team found that they could also use their DPGAs as a classification tool for small RNA molecules, which allowed for isolating certain types—such as those that were markers for certain types of cancer cells.
More information: Hui Lv et al, DNA-based programmable gate arrays for general-purpose DNA computing, Nature (2023). DOI: 10.1038/s41586-023-06484-9
Journal information: Nature
© 2023 Science X Network