This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Scientists study the biological process of after-ripening and dormancy release in rice seeds
After-ripening, an extension of seed maturation is pivotal for seed dormancy release, which has implications for germination and subsequent seedling robustness. The efficacy of this process varies with species traits, storage conditions, and environmental factors. Rice (Oryza sativa L.) is one of the most important food crops in China. Understanding the complex physiological and molecular processes in rice after-ripened seeds is paramount for enhancing agricultural productivity.
Seed Biology published a paper entitled, "Preliminary identification of the changes of physiological characteristics and transcripts in rice after-ripened seeds," on 25 April 2023.
In this study, researchers evaluated the process of after-ripening in Nipponbare (NPB) seeds, including germination percentage (GP), seedling percentage (SP), germination index (GI), and associated molecular and chemical changes at different time intervals (0, 28, and 56 days after ripening, DAR).
Freshly harvested NPB seeds (0 DAR) exhibited deep dormancy, only 10% GP, 6% SP and 0.3 GI. The GP, SP, and GI of NPB seeds were 98%, 98%, and 5.2 at 56 DAR.
Physiological results showed that at the early stage of after-ripening from 0 DAR to 28 DAR, abscisic acid (ABA) content significantly decreased, hydrogen peroxide (H2O2) levels significantly increased, superoxide dismutase (SOD) and α-amylase activities significantly raised, and bioactive gibberellin (GA1) content and ascorbate peroxidase (APX) activity showed no changes in NPB seeds.
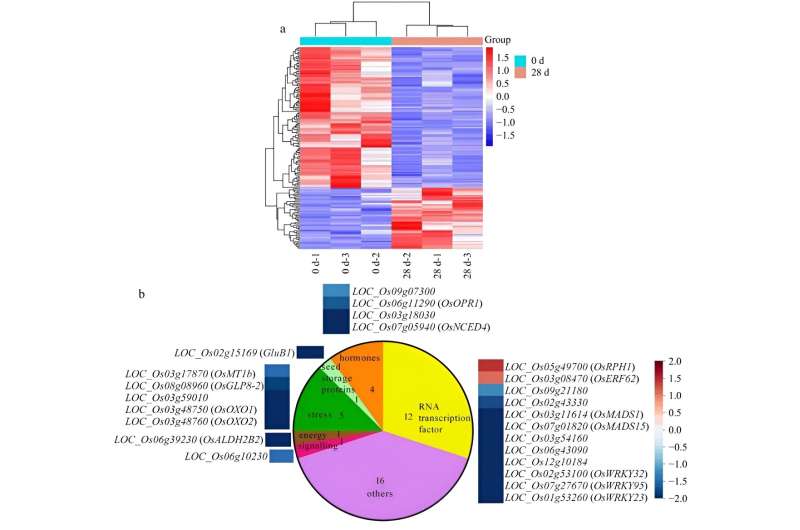
Using RNA-seq to investigate transcriptional changes between NPB dry seeds at 0 and 28 DAR, 149 differential transcripts were identified, encompassing key molecular pathways associated with seed dormancy. Among these, OsOxO1, OsNCED4, RSOsPR10 and OsPR10a, mainly occurred at the early stage of after-ripening through the verification of four other rice varieties in after-ripened seeds, suggesting the four transcripts play a key role in the regulation of rice seed after-ripening.
In conclusion, this study unravels multiple interactions of hormones, enzymes, and molecular pathways during after-ripening in rice seeds, providing a foundational framework for advanced seed biology research.
More information: Fengzhi Yuan et al, Preliminary identification of the changes of physiological characteristics and transcripts in rice after-ripened seeds, Seed Biology (2023). DOI: 10.48130/SeedBio-2023-0005
Provided by Maximum Academic Press