This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Mechanism underlying bacterial resistance to the antibiotic albicidin revealed
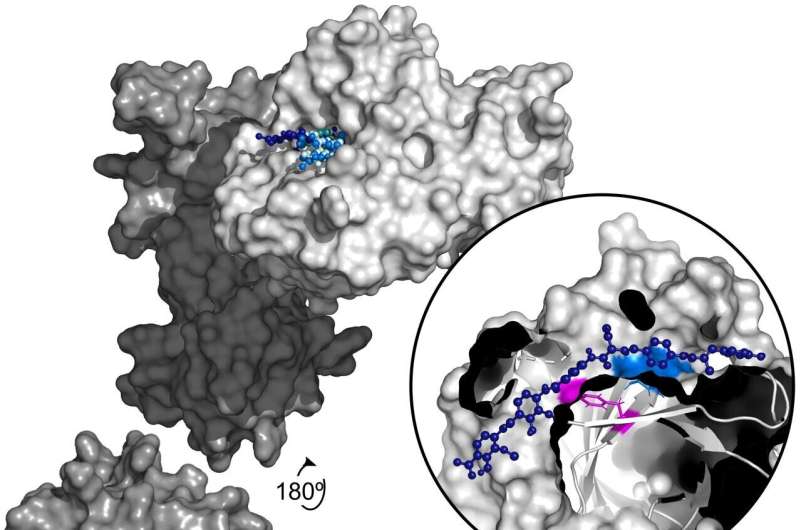
A new analysis shows that infectious bacteria exposed to the antibiotic albicidin rapidly develop up to a 1,000-fold increase in resistance via a gene amplification mechanism. Mareike Saathoff of Freie Universität Berlin, Germany, and colleagues present these findings in the open access journal PLOS Biology.
Bacterial resistance to antibiotics is a growing problem associated with millions of deaths around the world every year. Understanding how bacteria evolve resistance is key to developing more effective antibiotics and strategies for using them.
In recent years, albicidin has emerged as a promising antibiotic capable of killing a wide range of bacterial species by disrupting their DNA replication. Researchers are working to develop new albicidin-based medications; yet, despite its promise, some bacteria are able to develop resistance to albicidin.
To further investigate albicidin resistance mechanisms, Saathoff and colleagues conducted a suite of experiments employing a broad set of tools, including RNA sequencing, protein analysis, X-ray crystallography, and molecular modeling.
They found that two bacteria often associated with human infection—Salmonella typhimurium and Escherichia coli—develop resistance to albicidin when exposed to increasingly higher concentrations of the compound. Their analysis narrowed down the source of this resistance to an increase in the number of copies of a gene known as STM3175 (YgiV) in the bacterial cells, which is amplified in each new generation of cells as they multiply. STM3175 encodes a protein that interacts with albicidin in such a way that protects the bacteria from it.
Further experiments showed that the same albicidin-resistance mechanism is widespread among both pathogenic and harmless bacteria, including the microbes Vibrio vulnificus, which can infect wounds, and Pseudomonas aeruginosa, which can cause pneumonia and other infections. These findings could help inform the ongoing development of albicidin-based antibiotic strategies.
The authors add, "Our study reveals a gene duplication and amplification-based mechanism of a transcriptional regulator in Gram-negative bacteria, that mediates resistance to the peptide antibiotic albicidin."
More information: Saathoff M, Kosol S, Semmler T, Tedin K, Dimos N, Kupke J, et al. (2023) Gene amplifications cause high-level resistance against albicidin in gram-negative bacteria, PLoS Biology (2023). DOI: 10.1371/journal.pbio.3002186 , journals.plos.org/plosbiology/ … journal.pbio.3002186
Journal information: PLoS Biology
Provided by Public Library of Science