This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers discover mechanism by which cancer cells survive replication stress
Researchers from Karolinska Institutet have discovered a new molecular mechanism by which cancer cells safeguard themselves from oncogene-induced replication stress and propose a strategy to deactivate this protective mechanism. The study is published in the journal Molecular Cell.
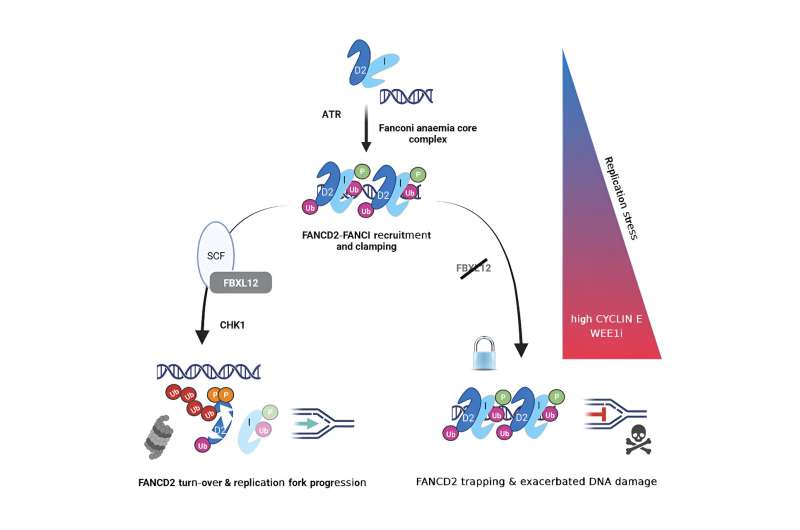
The study has revealed an unanticipated mechanism responsible for the removal of a pivotal DNA repair protein, FANCD2, at stopped replication forks. This enables the uninterrupted progression of DNA synthesis even in the presence of elevated oncogene-triggered replication stress, a characteristic inherent to cancer cells.
"We investigated how this process is regulated under conditions of replication stress and provided evidence that the SCF ubiquitin ligase receptor FBXL12 facilitates the proteasomal degradation of FANCD2. When FBXL12 is absent, FANCD2 becomes trapped at replication forks, impeding replication progression and survival specifically in cancer cells with high levels of replication stress," says Andrä Brunner, the study's first author from the Department of Cell and Molecular Biology, Karolinska Institutet
"Our cells face continuous DNA damage, especially during DNA synthesis governed by cyclin-dependent kinases (CDKs). Elevated CYCLIN E levels are associated with aggressive cancer types and unfavorable patient prognoses, posing inherent treatment challenges. From a clinical standpoint, CYCLIN E is not perceived as a feasible drug target. Our research expose an alternative strategy for eradicating cancer cells with CYCLIN E overexpression by targeting pathways that provide protection to the cancer genome."
"We initiated the study by carrying out a screen for genes regulating recovery of replication in cancer cells that overexpress CYCLIN E and identified FBXL12 as a prominent candidate. Employing high-resolution microscopy of individual DNA molecules and replication tracks, we next investigated how FBXL12 influences replication fork dynamics," Olle Sangfelt, the study's senior author says.
"After demonstrating that loss of FBXL12 blocked DNA synthesis, causing DNA damage and cell death in CYCLIN E overexpressing cells, we profiled the proteome for FBXL12 interacting proteins and identified FANCD2. Through in-depth molecular and biochemical analysis, we uncovered a crucial role of FBXL12-FANCD2 signaling in supporting the survival of cancer cells experiencing high levels of replication stress."
"Based on our findings, we propose that the disruption of FBXL12-FANCD2 signaling renders cells overexpressing CYCLIN E nonviable and hypothesize that targeting this pathway, alongside other cancer drugs inducing replication stress, may offer a feasible treatment avenue also for cancer types devoid of elevated replication stress levels. Our ongoing research delves into identifying and investigating compounds that disrupt FBXL12-FANCD2 signaling, to ultimately unleash the lethal potential of unchecked replication stress on cancer cells."
More information: Andrä Brunner et al, FBXL12 degrades FANCD2 to regulate replication recovery and promote cancer cell survival under conditions of replication stress, Molecular Cell (2023). DOI: 10.1016/j.molcel.2023.07.026
Journal information: Molecular Cell
Provided by Karolinska Institutet