This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study identifies characteristics specific to human brains
Researchers led by a team at UT Southwestern Medical Center have identified cellular and molecular features of the brain that set modern humans apart from their closest primate relatives and ancient human ancestors. The findings, published in Nature, offer new insights into human brain evolution.
"Most evolutionary studies on the human brain have focused on neurons because this cell type was thought to be responsible for our intelligence and enhanced cognitive abilities. This study gives us a renewed appreciation for other cells involved in brain function and the role they have played both in advancing cognition and our susceptibility to a number of cognitive diseases," said study leader Genevieve Konopka, Ph.D., Professor of Neuroscience and a member of the Peter O'Donnell Jr. Brain Institute at UT Southwestern.
Since ancient times, people have been curious about what gives humans abilities that other animals don't have, such as speech and language, Dr. Konopka explained. A range of previous studies have sought to answer this question by examining brain anatomy or performing genetic or molecular studies on whole brains or sections, experiments that provide a view of thousands of cells at a time.
Dr. Konopka and her colleagues theorized more could be gleaned from looking at brain characteristics at the cellular level, a feat only possible due to recent advances in technology. In this study, researchers in the Konopka lab, including lead author and O'Donnell Brain Institute Neural Scientist Training Program Fellow Emre Caglayan, B.S., together with colleagues at The George Washington University, Emory University, and the University of California, Santa Barbara, focused on Brodmann area 23 (BA23) in the posterior cingulate cortex. BA23 is also part of the default mode network—an interconnected complex of regions that remain active when the brain is in a state of wakeful rest—and has been implicated in schizophrenia.
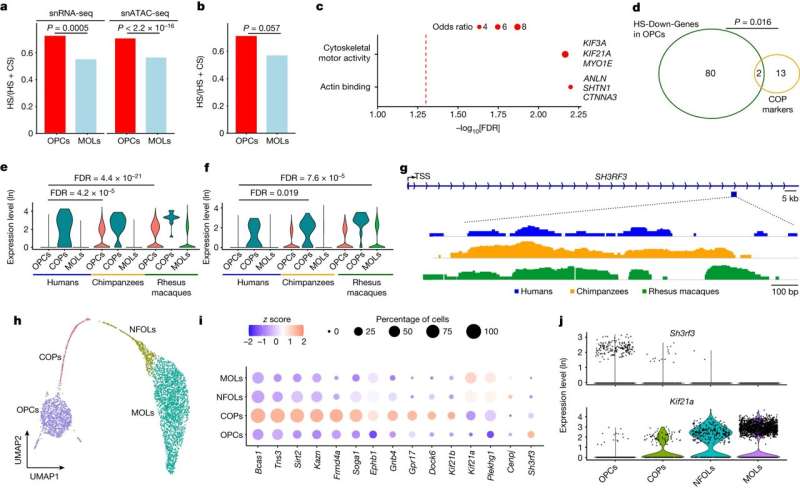
Rather than look at BA23 as a whole, the researchers used a relatively new technique called single nuclei RNA-sequencing to investigate what types of cells compose this area, comparing samples from humans, chimpanzees, and rhesus monkeys. They found that, in contrast to the nonhuman primates, humans have a far larger proportion of oligodendrocyte progenitor cells (OPCs), precursors to a type of cell known to provide support and insulation for neurons, and increasingly implicated in modulating brain circuitry. In addition, two subtypes of excitatory neurons—which share information through electrical impulses—showed increased expression in humans in the gene that makes FOXP2, a protein involved in brain development related to speech and language.
In another experiment, the researchers compared the DNA of modern humans with that of Neanderthals and Denisovans, ancient human relatives. They looked not only at differences in their genetic codes, but also whether these differences occurred in areas of the genome where cellular machinery regulates gene expression. Their search identified dozens of genes that functionally differ between humans and their ancient relatives, particularly in excitatory neurons in the upper layers of BA23, which could offer additional insight into human brain evolution in future studies.
Together, Dr. Konopka said, these findings offer a road map for understanding how human brains developed their unique abilities that set people apart from other species.
Dr. Konopka is a Jon Heighten Scholar in Autism Research and holds the Townsend Distinguished Chair in Research on Autism Spectrum Disorders. Other UTSW researchers who contributed to this study include postdoctoral researcher Yuxiang Liu, Ph.D., and graduate students Rachael Vollmer, B.S., and Emily Oh, B.S.
More information: Emre Caglayan et al, Molecular features driving cellular complexity of human brain evolution, Nature (2023). DOI: 10.1038/s41586-023-06338-4
Journal information: Nature
Provided by UT Southwestern Medical Center