This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers visualize activity of CRISPR genetic scissors in real time
When bacteria are attacked by a virus, they can defend themselves with a mechanism that fends off the genetic material introduced by the intruder. The key is CRISPR-Cas protein complexes. It is only in the last decade that their function for adaptive immunity in microorganisms has been discovered and elucidated.
With the help of an embedded RNA, the CRISPR complexes recognize a short sequence in the attacker's DNA. The mechanism of sequence recognition by RNA has since been used to selectively switch off and modify genes in any organism. This discovery revolutionized genetic engineering and was already honored in 2020 with the Nobel Prize in Chemistry awarded to Emmanuelle Charpentier and Jennifer A. Doudna.
Occasionally, however, CRISPR complexes also react to gene segments that differ slightly from the sequence specified by the RNA. This leads to undesirable side effects in medical applications. "The causes of this are not yet well understood, as the process could not be observed directly until now," says Dominik Kauert, who worked on the project as a Ph.D. student.
Nanoscale processes tracked in detail
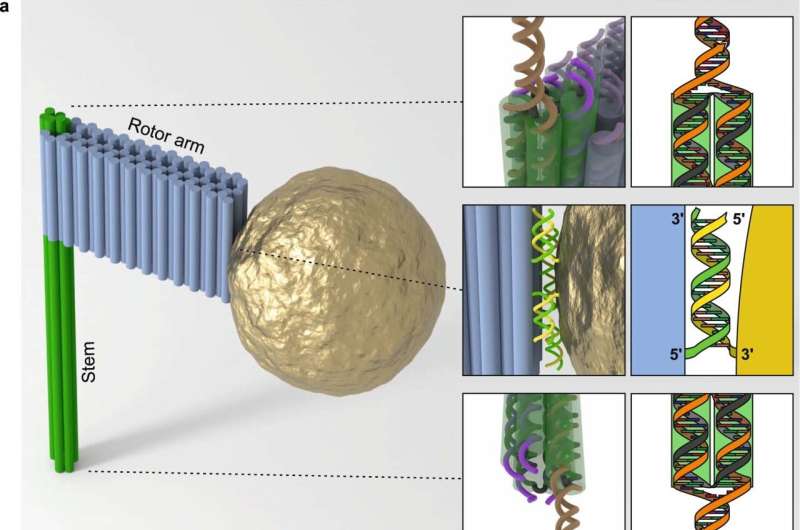
To better understand the recognition process, the team led by Professor Ralf Seidel and Dominik Kauert took advantage of the fact that the DNA double helix of the target sequence is unwound during recognition to enable base pairing with the RNA. "The central question of the project was therefore whether the unwinding of a piece of DNA that is only 10 nanometers (nm) long could be tracked in real time at all," says Kauert.
To observe the unwinding process in detail, the scientists had to make it visible to the microscope. To achieve this goal, the team drew on the achievements of DNA nanotechnology, which can be used to create any three-dimensional DNA nanostructure. Using this so-called DNA origami technique, the researchers constructed a 75 nm long DNA rotor arm with a gold nanoparticle attached to its end. In the experiment, the unwinding of the 2 nm thin and 10 nm long DNA sequence was transferred to the rotation of the gold nanoparticle along a circle with a diameter of 160 nm—this movement can be magnified and tracked using a special microscope setup.
With this new method, the researchers were able to observe the sequence recognition by the CRISPR Cascade complex almost base pair by base pair. Surprisingly, base pairing with the RNA is not energetically advantageous, meaning that the complex is only unstably bound during sequence recognition. Only when the entire sequence is recognized does stable binding occur and the DNA is subsequently destroyed. If it is the "wrong" target sequence, the process is aborted. The research is published in the journal Nature Structural & Molecular Biology.
Findings will help in selecting suitable RNA sequences
The fact that the recognition process sometimes produces incorrect results is due to its stochastic nature, i.e., to random molecular movements, as the researchers have now been able to demonstrate. "Sequence recognition is driven by thermal fluctuations in base pairing," says Kauert.
With the data obtained, it was possible to create a thermodynamic model of sequence recognition that describes the recognition of deviating sequence segments. In the future, this should allow better selection of RNA sequences that recognize only the desired target sequence, thus optimizing the precision of genetic manipulation.
As the designed nanorotors are universal in their suitability for measuring twists and torques in single molecules, they can also be used for other CRISPR-Cas complexes or biomolecules.
More information: Dominik J. Kauert et al, The energy landscape for R-loop formation by the CRISPR–Cas Cascade complex, Nature Structural & Molecular Biology (2023). DOI: 10.1038/s41594-023-01019-2
Journal information: Nature Structural & Molecular Biology
Provided by Leipzig University