This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers craft 'origami DNA' to control virus assembly
Griffith University researchers have played a key role in using DNA "origami" templates to control the way viruses are assembled.
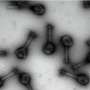
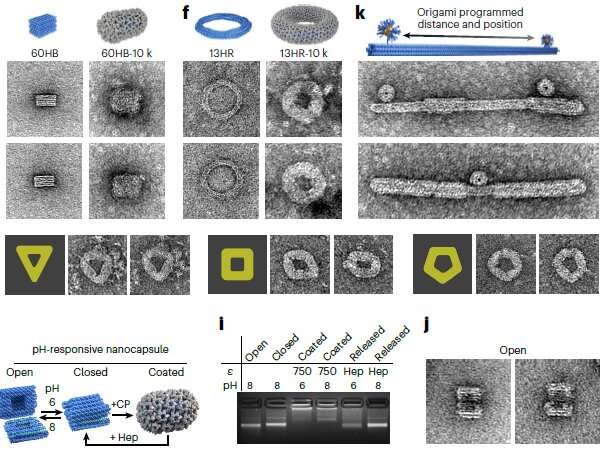
The global team behind the research, titled "DNA-origami-directed virus capsid polymorphism," published in Nature Nanotechnology, developed a way to direct the assembly of virus capsids—the protein shell of viruses—at physiological conditions in a precise and programmable manner.
Dr. Frank Sainsbury and Dr. Donna McNeale from the Griffith Institute for Drug Discovery were part of the research team and said forcing viruses to assemble onto DNA folded into different shapes "like origami" was a question that this project answered.
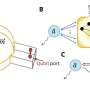
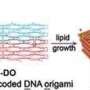
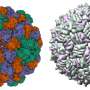
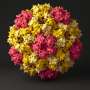
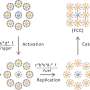
"We achieved control over the virus protein shape, size and topology by using user-defined DNA origami nanostructures as binding and assembly platforms, which became embedded within the capsid," Dr. Sainsbury said.
"The virus protein coatings could shield the encapsulated DNA origami from degradation.
"This activity is more like wrapping a present—the virus proteins deposit on top of the different shape that is defined by the DNA origami shape.
"And different virus proteins are like different wrapping paper, which would be relevant to different uses of the coated DNA origami."
Precise control over the size and shape of virus proteins would have advantages in the development of new vaccines and delivery systems.
"But current tools to control the assembly process in a programmable manner were elusive," Dr. McNeale said.
"Our approach is also not limited to a single type of virus capsid protein unit and can also be applied to RNA–DNA origami structures to pave way for next-generation cargo protection and targeting strategies."
Currently, Dr. Sainsbury and his team are working on gaining a more in-depth understanding of how different viruses self-assemble and how they can be used to encapsulate different cargoes.
This will allow them to design and modify further virus-like particles for a range of uses. For example, they discovered that one virus found in mice is able to carry protein cargoes through inhospitable environments and into a specific subcellular compartment in human cells.
"With the enormous existing design space among viruses that could be used as carriers, there is still much to learn from studying them. We'll continue to push the boundaries of how virus-like particles can assemble and what can be learned from using them as medicine transporters, vaccines and biochemical reaction vessels," Dr. Sainsbury said.
The next stage of the GRIDD team's research will use this approach to look at why viruses don't assemble into different shapes themselves.
More information: DNA-origami-directed virus capsid polymorphism, Nature Nanotechnology (2023). DOI: 10.1038/s41565-023-01443-x, https://www.nature.com/articles/s41565-023-01443-x
Journal information: Nature Nanotechnology
Provided by Griffith University