This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Computer scientists sequence cotton genome
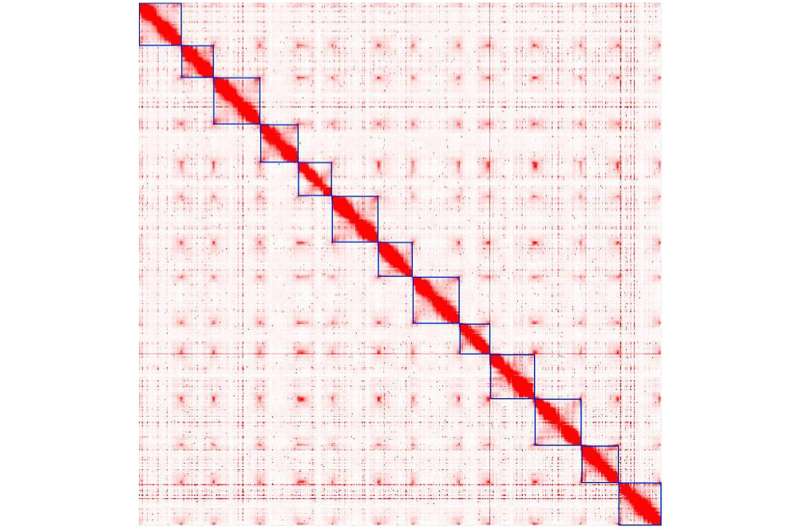
Cotton is the primary source of natural fiber on Earth, yet only four of 50 known species are suitable for textile production. Computer scientists at DePaul University applied a bioinformatics workflow to reconstruct one of the most complete genomes of a top cotton species, African domesticated Gossypium herbaceum cultivar Wagad. Experts say the results give scientists a more complete picture of how wild cotton was domesticated over time and may help to strengthen and protect the crop for farmers in the U.S., Africa and beyond.
The findings are published in the journal G3 Genes|Genomes|Genetics. Thiru Ramaraj, assistant professor of computer science in DePaul's Jarvis College of Computing and Digital Media, is lead author on the publication. Leaps in technological advancement in the past decade made it possible for Ramaraj to analyze the genome in his Chicago lab.
"The power of this technology is it allows us to create high-quality genomes that supply a level of detail that simply wasn't possible before," says Ramaraj, who specializes in bioinformatics. "This opens up the possibility for more researchers to sequence many crops that are important to the global economy and to feeding the population."
The work is part of a collaboration that includes Jonathan Wendel, distinguished professor in the Department of Ecology, Evolution, and Organismal Biology at Iowa State University; and Joshua Udall, research leader for the Crop Germplasm Research Unit at the U.S. Department of Agriculture Agricultural Research Service. According to Udall, Wagad cotton is a diploid strain grown predominantly in African countries. "This has the potential to provide a genetic map that could improve their cotton crop," Udall said.
Advanced computational methodologies bring forward genome
The team's work began with crunching DNA sequence data. They began reconstructing the Wagad genome by assembling high quality long DNA sequence data generated using Pacific Biosciences sequencing technology. As a next step, whole genome maps from Bionano genomics were used to order and orient the initial assembly. Lastly Hi-C sequence data from Phase genomics were used to construct chromosome level genome.
Ramaraj then turned to Azalea Mendoza, a graduate student in computer science who also holds a bachelor's degree in environmental studies from DePaul. "Azalea had the biology background and knowledge to dive into this research," Ramaraj said.
Mendoza began by researching the history of cotton to zoom out and understand "the big picture." No matter where cotton is grown, it's primarily used for fiber. Using comparative genomics, she looked for variations against its closet relative and to an outgroup. Mendoza also delved into annotated genes and noted their functions. "As we were studying the regions of the genome, we found many genes that were related to the content of fiber," Mendoza says. "It was incredible to see the real-life application of the work."
Protecting crops in the U.S. and beyond
The impact of cotton genomics on U.S. agriculture and economy are clear to Udall, who has worked with Ramaraj since 2015. Udall leads the Crop Germplasm Research Unit and examines some of the 10,000 accessions of various species that the USDA holds in its repository. Their goal is to maintain the country's genetic food and feed security, and part of that is understanding the resilience and weaknesses of crops from around the world.
"When new diseases come to the U.S., or there's new invasive pests, one of the first things we do is screened the genetic diversity of cotton to see if any of the previous varieties are resistant to it," Udall says. This can give farmers a chance to cross breed those genes and improve modern varieties of cotton, potentially avoiding catastrophic loss of agriculture.
Udall relies on computational biologists like Ramaraj to further this work. While the cost of sequencing genomes has come down, this study still took nearly two years of work across disciplines. "This is a good step in identifying future cotton genomes to sequence," Udall says.
Ramaraj hopes the project will inspire other faculty and student collaborators to approach CDM with ideas for bioinformatics projects. For Mendoza, now an alumna working as a data analyst, the experience working in bioinformatics at DePaul is inspiring her career goals.
"I love research and work that helps me grow on multiple levels," Mendoza says. "This is the kind of work that is going to affect humans and sustainability into the future."
More information: Thiruvarangan Ramaraj et al, The Gossypium herbaceum L. Wagad genome as a resource for understanding cotton domestication, G3 Genes|Genomes|Genetics (2022). DOI: 10.1093/g3journal/jkac308
Journal information: G3 Genes|Genomes|Genetics
Provided by DePaul University





















