May 18, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Optimized prime editing alters genes of living mice, marking major advance
Harvard University researchers have improved a gene editing process for studying and treating genetic disorders. The prime editing method is effective in human cells, targeting single nucleotide variants with the ability to precisely correct pathogenic mutations or install more protective variants. While success in the lab has been promising, translating these effects into living systems where the therapeutic potential could be unleashed has proven tricky.
In a new paper, "Efficient prime editing in mouse brain, liver and heart with dual AAVs," published in Nature Biotechnology, the researchers detail the identification of bottlenecks limiting the effectiveness of adeno-associated virus (AAV)-mediated prime editing in vivo and the development of vectors with increased prime editing expression, RNA stability and modulation of DNA repair.
Prime editing is an ingenious strategy of editing that, in its basic configuration, utilizes the Cas9 from CRISPR and attaches an RNA scaffolding and a reverse transcriptase enzyme to create an accurate, precise editing system.
One of the advantages of prime editing is that the system does cause double-strand breaks, instead nicking one side to open the DNA. The approach minimizes unwanted insertions and deletions (indels) that can take place with other editing systems.
The method works well enough in a lab environment where you can place the editor near its target, but a living body has the added difficulty of needing to navigate a complex environment. In the current round of optimization, the researchers looked to deliver the editing system to the correct location in a living body.
The currently favored method of targeted gene delivery is adeno-associated virus (AAV) vectors, which can carry a small payload to a specific locus on the chromosome. That payload capacity is, unfortunately, too small to contain the prime editing mechanism.
Some assembly required
The solution to overcoming the vector capacity problem was to break the prime editor into smaller pieces that could fit within the AAV cargo limit and would self-assemble when in proximity to each other. With the vector situation somewhat solved, the team moved on to the next series of obstacles.
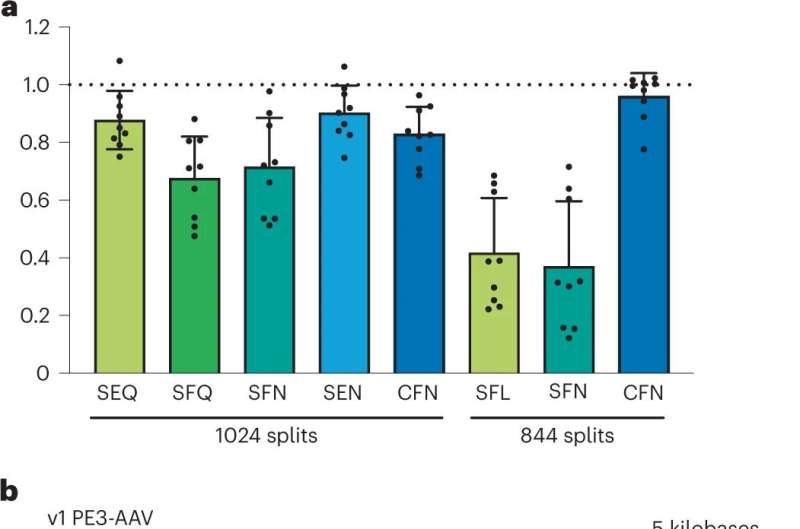
In a series of efforts, the teams made use of a woodchuck hepatitis virus regulatory element, reinforced the RNA scaffolding, switched promoters, ran out of room, added a third AAV vector, optimized the reverse transcriptase enzyme, modified Cas9 with a mutation, made sure everything was working, then removed any elements they could to downsize it back to a two AAV vector system.
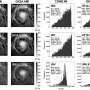
The resulting prototype for optimized prime editing, v3em PE-AAVs, was tested in mice. Therapeutically relevant levels of prime editing in the mouse brain (up to 42% efficiency), liver (up to 46%) and heart (up to 11%) were achieved. In vivo, prime editing resulted in no detected off-target editing. As a proof of concept, the gene change in the brain was specific to Alzheimer's disease resistance and a gene related to cholesterol-induced coronary artery disease was targeted in the liver.
Kind of a big deal
These levels of correcting mutations and rewriting variants summon up a host of often undeserved phraseologies in scientific circles as it truly is a groundbreaking, monumental, landmark, breakthrough, game-changing, unprecedented, a major milestone and, yes, even a paradigm-shifting moment.
The genetic mechanisms discovered in the past few decades and all of the knowledge accumulated on causal relationships between cellular machinery and disease pathologies imply that we will one day be able to repair or replace the faulty components of our genome.
The latest version of optimized prime editing is not quite ready to proofread the human genome for errors, but it is the closest thing on the planet to a future where we can change the genetic cards we were dealt at birth. A future where cancer risk factors can be eliminated, neurological disorders routinely prevented or reversed—a future where a long list of well-known diseases are simply forgotten. This is a future that is in the testing phase now.
More information: Jessie R. Davis et al, Efficient prime editing in mouse brain, liver and heart with dual AAVs, Nature Biotechnology (2023). DOI: 10.1038/s41587-023-01758-z
Journal information: Nature Biotechnology
© 2023 Science X Network