This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A simple paper test could offer early cancer diagnosis
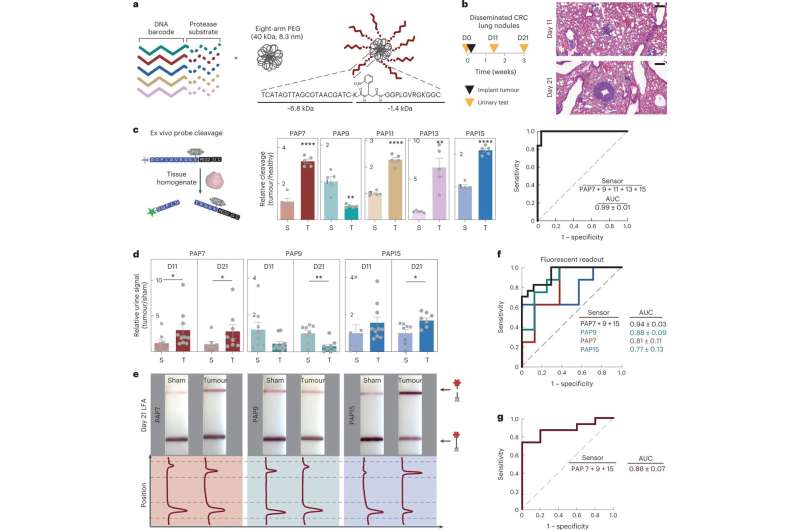
MIT engineers have designed a new nanoparticle sensor that could enable early diagnosis of cancer with a simple urine test. The sensors, which can detect many different cancerous proteins, could also be used to distinguish the type of a tumor or how it is responding to treatment.
The nanoparticles are designed so that when they encounter a tumor, they shed short sequences of DNA that are excreted in the urine. Analyzing these DNA "barcodes" can reveal distinguishing features of a particular patient's tumor. The researchers designed their test so that it can be performed using a strip of paper, similar to an at-home COVID test, which they hope could make it affordable and accessible to as many patients as possible.
"We are trying to innovate in a context of making technology available to low- and middle-resource settings. Putting this diagnostic on paper is part of our goal of democratizing diagnostics and creating inexpensive technologies that can give you a fast answer at the point of care," says Sangeeta Bhatia, the John and Dorothy Wilson Professor of Health Sciences and Technology and of Electrical Engineering and Computer Science at MIT and a member of MIT's Koch Institute for Integrative Cancer Research and Institute for Medical Engineering and Science.
In tests in mice, the researchers showed that they could use the sensors to detect the activity of five different enzymes that are expressed in tumors. They also showed that their approach could be scaled up to distinguish at least 46 different DNA barcodes in a single sample, using a microfluidic device to analyze the samples.
Bhatia is the senior author of the paper, which appears today in Nature Nanotechnology. Liangliang Hao, a former MIT research scientist who is now an assistant professor of biomedical engineering at Boston University, is the lead author of the study.
DNA barcodes
For several years, Bhatia's lab has been developing "synthetic biomarkers" that could be used to diagnose cancer. This work builds on the concept of detecting cancer biomarkers, such as proteins or circulating tumor cells, in a patient's blood sample. These naturally occurring biomarkers are so rare that it's nearly impossible to find them, especially at an early stage, but synthetic biomarkers can be used amplify smaller-scale changes that occur within small tumors.
In previous work, Bhatia created nanoparticles that can detect the activity of enzymes called proteases, which help cancer cells to escape their original locations, or settle into new ones, by cutting through proteins of the extracellular matrix. The nanoparticles are coated with peptides that are cleaved by different proteases, and once these peptides are released into the bloodstream, they can then be concentrated and more easily detected in a urine sample.
The original peptide biomarkers were designed to be detected based on small engineered variations in their mass, using a mass spectrometer. This kind of equipment might not be available in low-resource settings, so the researchers set out to develop sensors that could be analyzed more easily and affordably, using DNA barcodes that can be read using CRISPR technology.
For this approach to work, the researchers had to use a chemical modification called phosphorothioate to protect the circulating DNA reporter barcodes from being broken down in the blood. This modification has already been used to improve the stability of modern RNA vaccines, allowing them to survive longer in the body.
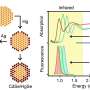
Similar to the peptide reporters, each DNA barcode is attached to a nanoparticle by a linker that can be cleaved by a specific protease. If that protease is present, the DNA molecule is released and free to circulate, eventually ending up in the urine. For this study, the researchers used two different types of nanoparticles: one, a particle made from polymers that have been FDA-approved for use in humans, and the other a "nanobody"—an antibody fragment that can be designed to accumulate at a tumor site.
Once the sensors are secreted in the urine, the sample can be analyzed using a paper strip that recognizes a reporter that is activated by a CRISPR enzyme called Cas12a. When a particular DNA barcode is present in the sample, Cas12a amplifies the signal so that it can be seen as a dark strip on a paper test.
The particles can be designed to carry many different DNA barcodes, each of which detects a different type of protease activity, which allows for "multiplexed" sensing. Using a larger number of sensors provides a boost in both sensitivity and specificity, allowing the test to more easily distinguish between tumor types.
Disease signatures
In tests in mice, the researchers showed that a panel of five DNA barcodes could accurately distinguish tumors that first arose in the lungs from tumors formed by colorectal cancer cells that had metastasized to the lungs.
"Our goal here is to build up disease signatures and to see whether we can use these barcoded panels not only read out a disease but also to classify a disease or distinguish different cancer types," Hao says.
For use in humans, the researchers expect that they may need to use more than five barcodes because there is so much variety between patients' tumors. To help reach that goal, they worked with researchers at the Broad Institute of MIT and Harvard led by Harvard University Professor Pardis Sabeti, to create a microfluidic chip that can be used to read up to 46 different DNA barcodes from one sample.
This kind of testing could be used not only for detecting cancer, but also for measuring how well a patient's tumor responds to treatment and whether it has recurred after treatment. The researchers are now working on further developing the particles with the goal of testing them in humans. Glympse Bio, a company co-founded by Bhatia, has performed phase 1 clinical trials of an earlier version of the urinary diagnostic particles and found them to be safe in patients.
More information: Liangliang Hao et al, CRISPR-Cas-amplified urinary biomarkers for multiplexed and portable cancer diagnostics, Nature Nanotechnology (2023). DOI: 10.1038/s41565-023-01372-9
Journal information: Nature Nanotechnology
Provided by Massachusetts Institute of Technology
This story is republished courtesy of MIT News (web.mit.edu/newsoffice/), a popular site that covers news about MIT research, innovation and teaching.