This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
How AI might speed up the discovery of new drugs
Artificial intelligence can generate poems and essays, create responsive game characters, analyze vast amounts of data and detect patterns that the human eye might miss. Imagine what AI could do for drug discovery, traditionally a time-consuming, expensive process from the bench to the bedside.
Experts see great promise in a complementary approach using AI and structure-based drug discovery, a computational method that relies on knowledge of 3D structures of biological targets.
We recently caught up with Vsevolod "Seva" Katritch, associate professor of quantitative and computational biology and chemistry at the USC Dornsife College of Letters, Arts and Sciences and the USC Michelson Center for Convergent Bioscience. Katritch is the co-director of the Center for New Technologies in Drug Discovery and Development (CNT3D) at the USC Michelson Center and the lead author of a new review paper published in Nature. The paper, co-authored by USC research scientist Anastasiia Sadybekov, describes how computational approaches will streamline drug discovery.
We're on the cusp of major advances in drug discovery. What brings us to this moment?
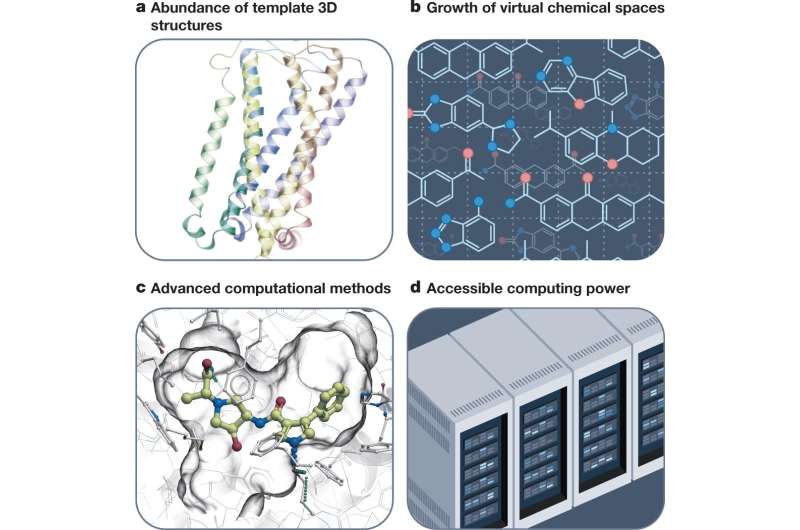
There has been a seismic shift in computational drug discovery in the last few years: an explosion of data availability on clinically relevant, human-protein structures—and molecules that bind them, enormous chemical libraries of drug-like molecules, almost unlimited computing power and new, more efficient computational methods. The newest excitement is about AI-based drug discovery, but what's even more powerful is a combination of AI and structure-based drug discovery, with both approaches synergistically complementing each other.
How has drug discovery been done in the past?
Traditional drug discovery is mostly a trial-and-error venture. It's slow and expensive, taking an average of 15 years and $2 billion. There's a high attrition rate at every step, from target selection to lead optimization. The most opportunities for time and cost savings reside in the earlier discovery and preclinical stages.
What takes place in the early stage?
Let's use a lock-and-key analogy. The target receptor is the lock, and the drug that blocks or activates this receptor is a key for this lock. (Of course, the caveat is that in biology nothing is black or white, so some of the working keys switch the lock better than others, and lock is a bit malleable too.)
Here's an example. Lipitor, the bestselling drug of all time, targets an enzyme involved in the synthesis of cholesterol in the liver. A receptor on the enzyme is the lock. Lipitor is the key, fitting into the lock and blocking the activity of the enzyme, triggering a series of events that decrease blood levels of bad cholesterol.
Now, computational approaches allow us to digitally model many billions and even trillions of virtual keys and predict which ones are likely to be good keys. Only a few dozen of the best candidate keys are chemically synthesized and tested.
This sounds much more efficient
If the model is good, this process yields better results than traditional trial-and-error testing of millions of random keys. This reduces the physical requirements for synthesis of compounds and testing them more than thousandsfold, while often arriving at better results, as demonstrated by our work and work of many other groups working in this field.
Can you explain the difference between the two main computational approaches, structure-based and AI-based?
Following the lock-and-key analogy, the structure-based approach takes advantage of our detailed understanding of the lock's structure. If the 3D, physical structure of the lock is known, we can use virtual methods to predict the structure of a key that matches the lock.
The machine learning, or AI-based approach, works best when many keys are already known for our target lock or other similar locks. AI can then analyze this mixture of similar locks and keys and predict the keys that are most likely to fit our target. It does not need exact knowledge of the lock structure, but it needs a large collection of relevant keys.
Thus, the structure-based and AI-based approaches are applicable in different cases and complement each other.
Are there any computational limits to this process?
When testing billions and trillions of virtual compounds on cloud computers, computational costs themselves can become a bottleneck. A modular, giga-scale screening technology allows us to speed up and reduce cost dramatically by virtually predicting good parts of the key, combine them together, sort of building the key from several parts. For a 10 billion-compound library, this drops the computational costs from millions of dollars to hundreds, and it allows further scale-ups to trillions of compounds.
More information: Anastasiia V. Sadybekov et al, Computational approaches streamlining drug discovery, Nature (2023). DOI: 10.1038/s41586-023-05905-z
Journal information: Nature
Provided by University of Southern California