This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Physicists investigate the dynamics of active protein droplets in cells
The phase separation of oil and water is a classic example of a process ubiquitous in nature, in which mixtures separate into their constituent parts. Scientists have also repeatedly identified biomolecular droplets in cells that originate from the phase-separation of proteins and nucleic acids. These droplets play various vital roles in cells where, for example, they create a local environment for chemical reactions, and are responsible for the midcell localization of proteins during cell division.
A team led by LMU physicist Professor Erwin Frey has now investigated the behavior of enzymatic droplets and predicted that they can show directed self-propulsion in the cell. The study is published in the journal Physical Review Letters.
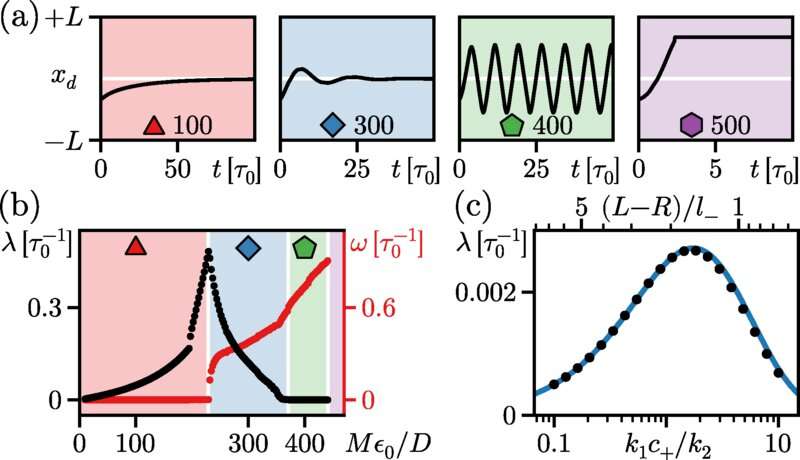
In a thermodynamic system that is in thermal equilibrium, large droplets grow at the expense of smaller droplets, thus rendering a state with many droplets unstable. In a cell, by contrast, several studies have demonstrated droplets are often driven out of equilibrium by influx of energy. "This leads to a wealth of novel phenomena," says Frey. With his team, he investigated the mechanisms of droplets made up of enzymes that catalyze chemical reactions between proteins. Using a minimal model, the researchers were able to show that these enzymatic droplets can direct self-propulsion and that, in closed domains, they move to the center in a directed manner.
"This property resembles the mechanism by which protein droplets find the cell middle in prokaryotes (bacteria) and can control correct cell division," says Frey. Under other conditions, however, the droplets, depending on their size, can also divide, resulting in a stable coexistence of multiple droplets. "This stands in contrast to the dynamics that one would expect in thermal equilibrium," says Frey. "Our results underscore the role of chemical reactions in droplet formation, representing a broad and very active research domain."
More information: Leonardo Demarchi et al, Enzyme-Enriched Condensates Show Self-Propulsion, Positioning, and Coexistence, Physical Review Letters (2023). DOI: 10.1103/PhysRevLett.130.128401
Journal information: Physical Review Letters
Provided by Ludwig Maximilian University of Munich