This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Research team identifies new mtDNA editing tool
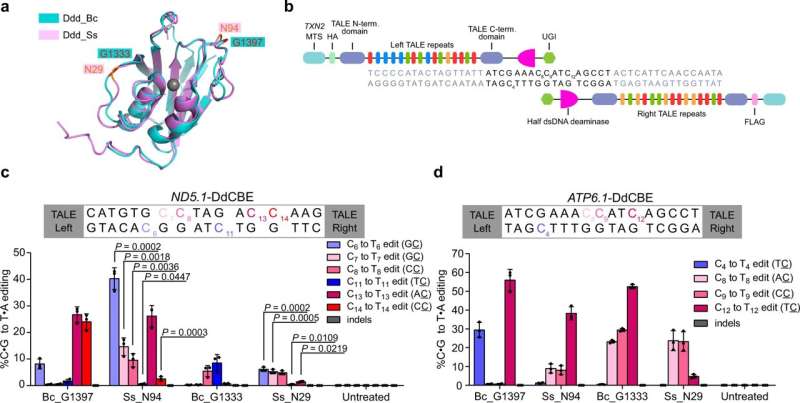
A team of researchers from the Wang Yangming Laboratory at Peking University's College of Future Technology has recently identified a DddA homolog from Simiaoa sunii (Ddd_Ss) that can efficiently deaminate cytosine in DC context in double-stranded DNA (dsDNA). Their findings were published on February 16, 2023 in Nature Communications, titled "DddA homolog search and engineering expand sequence compatibility of mitochondrial base editing."
Base editing is a precise way to introduce mutations to a target DNA sequence, making it useful for studying genes and regulatory elements, disease modeling, and developing treatments for genetic diseases. While base editing for nuclear DNA has been achieved through clustered regularly interspaced short palindromic repeats (CRISPR) derived base editors, the same approach is not applicable for mitochondrial DNA (mtDNA) editing because of the difficulty in delivering guide RNAs into mitochondria.
Researchers have been using transcription activator-like-effector (TALE) derived base editors DdCBEs and TALEDs to catalyze C-to-T and A-to-G editing in mtDNA. These methods rely on DddAtox, a dsDNA deaminase from the bacterium Burkholderia cenocepacia (Ddd_Bc). However, the original Ddd_Bc requires a strict TC sequence context, which makes it unsuitable for GC context targets. Furthermore, it is unknown whether more dsDNA deaminases can be found in other species.
To address these problems, the researchers in this study identified and engineered dsDNA deaminase homologs that could be used to develop highly efficient base editing tools. They discovered a dsDNA deaminase from Simiaoa sunii (named after Sun Simiao, a great medical scientist of China in the Tang Dynasty (618–907) that demonstrated high activity and broad sequence compatibility. Based on this discovery, they developed highly efficient base editing tools that could introduce mutations at multiple mtDNA loci, including disease-related mutations in previously inaccessible GC context. They also improved the activity and sequence compatibility of DdCBE_Bc by introducing a single amino acid substitution from Ddd_Ss.
The researchers' results showed that the C-terminus of Ddd_Bc contains two SPKK-related peptide motifs that prefer the binding of A/T-rich DNA sequences in the minor groove of dsDNA. Deletion of these motifs eliminated the dsDNA deaminase activity of Ddd_Bc. However, the addition of an AT-hook, which has a similar DNA binding property as SPKK-related motifs, restored the deaminase activity of truncated Ddd_Bc. Mutation of prolines to valine or asparagine in C-terminal peptide reduced or completely abolished the deaminase activity of Ddd_Bc, indicating the importance of the SPKK-related motif in the C-terminus of Ddd_Bc for its dsDNA deamination activity.
This study expands the sequence compatibility of mitochondrial base editors, but more sophisticated designs will be needed in the future to achieve efficient and highly specific editing.
Mi Li, a Ph.D. student at the College of Future Technology of Peking University, and Ming Shi, a Ph.D. student at the Peking University-Tsinghua University Life Science Joint Center, are the co-first authors of the paper. The study was guided and assisted by the laboratories of Yi Chengqi, Gao Ning, Wei Wensheng, Cheng Heping, Wang Xianhua, and Chen Liangyi at Peking University. A patent application for the newly discovered double-stranded deaminase-related application has also been filed.
More information: Li Mi et al, DddA homolog search and engineering expand sequence compatibility of mitochondrial base editing, Nature Communications (2023). DOI: 10.1038/s41467-023-36600-2
Journal information: Nature Communications
Provided by Peking University





















