This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Identifying intestinal microbiota bacteria that protect against antibiotic-resistant pathogens
A study by the "Microbiota, Infection and Inflammation" research group at the Foundation for the Promotion of Health and Biomedical Research of Valencia Region (Fisabio), an agency of the Conselleria de Sanitat Universal i Salut Pública, has identified the ability of five bacterial strains of intestinal microbiota to restrict the colonization of bacteria resistant to multiple antibiotics.
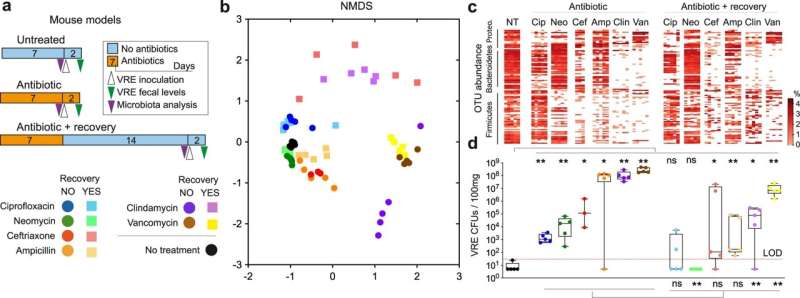
This study, led by Dr. Carles Úbeda and published in Nature Communications, has revealed that in animal models, the consortium of bacterial strains of the genera Alistipes, Barnesiella, Olsenella, Oscillibacter and Flavonifractor—which are naturally present in the microbiota intestine—depletes the nutrients necessary for the growth of bacteria of the genus Enterococcus. These pathogens, which are multi-resistant to antibiotics, are especially affected by the loss of a type of sugar called fructose, which is commonly found in our diet.
In this way, the nutritional deficiency that pathogens encounter prevents their optimal growth and consequently protects the body against infection. "Using models with mice, we have shown that the administration of these commensal bacteria decreases the ability of the pathogen to colonize the intestine, a key step for the development of infection and transmission between patients," explained Dr. Úbeda.
These types of pathogens are among the third and fourth most prevalent causes of infections in hospitalized patients worldwide and can cause lethal outcomes due to their resistance to most currently available antibiotics, making treatment difficult. For this reason, it is among the highest priority multi-resistant pathogens for which new therapies must be developed, according to the World Health Organization (WHO).
To conduct the study, the research group of the Genomics and Health Area has applied novel techniques of mass-sequencing of bacterial DNA (metagenomics) and RNA (transcriptomics), as well as the analysis of substances called metabolites (metabolomics) present in the gastrointestinal tract.
This study could lead to new non-antibiotic-based strategies to prevent infections caused by this multi-resistant pathogen. "This is a significant discovery, because antibiotic resistance is one of the most important public health problems facing society today," concluded the researcher.
More information: Sandrine Isaac et al, Microbiome-mediated fructose depletion restricts murine gut colonization by vancomycin-resistant Enterococcus, Nature Communications (2022). DOI: 10.1038/s41467-022-35380-5
Journal information: Nature Communications
Provided by Foundation for the Promotion of Health and Biomedical Research in the Valencian Region (FISABIO)