This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Gone fishing: Highly accurate test for common respiratory viruses uses DNA as 'bait'

Cambridge researchers have developed a new test that "fishes" for multiple respiratory viruses at once using single strands of DNA as bait and gives highly accurate results in under an hour.
The test uses DNA "nanobait" to detect the most common respiratory viruses—including influenza, rhinovirus, RSV and COVID-19—at the same time. In comparison, PCR (polymerase chain reaction) tests, while highly specific and highly accurate, can only test for a single virus at a time and take several hours to return a result.
While many common respiratory viruses have similar symptoms, they require different treatments. By testing for multiple viruses at once, the researchers say their test will ensure patients get the right treatment quickly and could also reduce the unwarranted use of antibiotics.
In addition, the tests can be used in any setting, and can be easily modified to detect different bacteria and viruses, including potential new variants of SARS-CoV-2, the virus which causes COVID-19. The results are reported in the journal Nature Nanotechnology.
The winter cold, flu and RSV season has arrived in the northern hemisphere, and health care workers must make quick decisions about treatment when patients show up in their hospital or clinic.
"Many respiratory viruses have similar symptoms but require different treatments: we wanted to see if we could search for multiple viruses in parallel," said Filip Bošković from Cambridge's Cavendish Laboratory, the paper's first author. "According to the World Health Organization, respiratory viruses are the cause of death for 20% of children who die under the age of five. If you could come up with a test that could detect multiple viruses quickly and accurately, it could make a huge difference."
For Bošković, the research is also personal: as a young child, he was in hospital for almost a month with a high fever. Doctors could not figure out the cause of his illness until a PCR machine became available.
"Good diagnostics are the key to good treatments," said Bošković, who is a Ph.D. student at St John's College, Cambridge. "People show up at hospital in need of treatment and they might be carrying multiple different viruses, but unless you can discriminate between different viruses, there is a risk patients could receive incorrect treatment."
PCR tests are powerful, sensitive and accurate, but they require a piece of genome to be copied millions of times, which takes several hours.
The Cambridge researchers wanted to develop a test that uses RNA to detect viruses directly, without the need to copy the genome, but with high enough sensitivity to be useful in a health care setting.
"For patients, we know that rapid diagnosis improves their outcome, so being able to detect the infectious agent quickly could save their life," said co-author Professor Stephen Baker, from the Cambridge Institute of Therapeutic Immunology and Infectious Disease. "For health care workers, such a test could be used anywhere, in the UK or in any low- or middle-income setting, which helps ensure patients get the correct treatment quickly and reduce the use of unwarranted antibiotics."
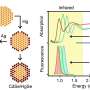
The researchers based their test on structures built from double strands of DNA with overhanging single strands. These single strands are the "bait": they are programmed to "fish" for specific regions in the RNA of target viruses. The nanobaits are then passed through very tiny holes called nanopores. Nanopore sensing is like a ticker tape reader that transforms molecular structures into digital information in milliseconds. The structure of each nanobait reveals the target virus or its variant.
The researchers showed that the test can easily be reprogrammed to discriminate between viral variants, including variants of the virus that causes COVID-19. The approach enables near 100% specificity due to the precision of the programmable nanobait structures.
"This work elegantly uses new technology to solve multiple current limitations in one go," said Baker. "One of the things we struggle with most is the rapid and accurate identification of the organisms causing the infection. This technology is a potential game changer; a rapid, low-cost diagnostic platform that is simple and can be used anywhere on any sample."
A patent on the technology has been filed by Cambridge Enterprise, the University's commercialization arm, and co-author Professor Ulrich Keyser has co-founded a company, Cambridge Nucleomics, focused on RNA detection with single-molecule precision.
"Nanobait is based on DNA nanotechnology and will allow for many more exciting applications in the future," said Keyser, who is based at the Cavendish Laboratory. "For commercial applications and roll-out to the public we will have to convert our nanopore platform into a hand-held device."
"Bringing together researchers from medicine, physics, engineering and chemistry helped us come up with a truly meaningful solution to a difficult problem," said Bošković, who received a 2022 Ph.D. award from Cambridge Society for Applied Research for this work.
More information: Ulrich Keyser, Simultaneous identification of viruses and viral variants with programmable DNA nanobait, Nature Nanotechnology (2023). DOI: 10.1038/s41565-022-01287-x. www.nature.com/articles/s41565-022-01287-x
Journal information: Nature Nanotechnology
Provided by University of Cambridge