September 19, 2022 report
Using AI to identify genomic tradeoffs between types of mutations
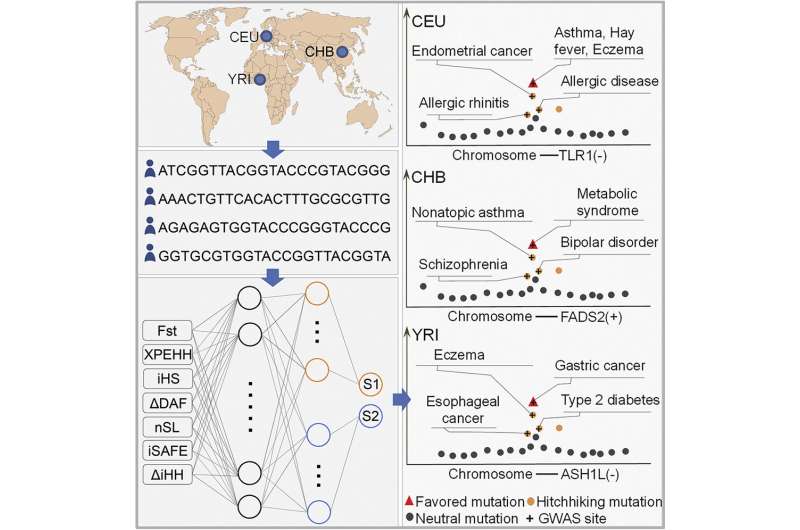
A team of researchers at Southern Medical University has developed an AI application to help identify genomic tradeoffs between different types of mutations that occurred as humans evolved. In their paper published in the journal Cell Reports, the group describes how they used data from currently existing genome-wide associations to teach their system and what it showed when exposed to new data.
Prior research has shown that as creatures evolve, mutations occur. Mutations that remain in the genome are what lead a creature to evolve. Prior research has also shown that some mutations lead to direct benefits, such as the ability to process certain foods, which allows a creature to exist in a new environment. Other mutations, on the other hand, sometimes just go along for the ride. They do not necessarily provide any benefits but remain in the genome accidentally or because of their proximity to genes that do provide a benefit.
Scientists studying the genome have long desired a tool that could be used to determine which mutations in the human genome were favored and which were merely hitchhikers. In this new effort, the researchers have developed such a tool, though it is still not clear how useful it truly is.
Called DeepFavored, the tool was created by developing a deep learning AI system that was fed data from existing genome-wide association studies to learn from the experiences of other researchers working on prior specific efforts. Those included by the team were narrowed down to alleles related to diet and other metabolic activities, and also mutations that allowed for dealing with variations in climate—the focus was on the unique ability of humans to adapt to so many different parts of the planet. The team then ran the tool on three separate populations and found what they describe as examples of hitchhiking mutations that have led to disease susceptibility.
In testing their new tool, the researchers also found what they describe as favored mutations among the three populations they tested. They suggest their overall findings indicate that their tool is capable of finding evidence for mutational trade-offs in the human genome. They also compared results from DeepFavored with two other algorithms created by other teams and found that it outperformed both.
More information: Ji Tang et al, Uncovering the extensive trade-off between adaptive evolution and disease susceptibility, Cell Reports (2022). DOI: 10.1016/j.celrep.2022.111351
Journal information: Cell Reports
© 2022 Science X Network