Organ-development discovery could boost battle against cancer
A new discovery from the University of Virginia School of Medicine has shed light on how our digestive tract, lungs and liver form, and that finding could have important implications for our understanding of cancer.
During development in mammals, the stomach, colon, intestines, pancreas, liver, lungs, esophagus, pharynx (throat) and thyroid all form from what is called a "primitive gut tube." But scientists have been uncertain exactly what prompts the indistinguishable cells in the gut tube to turn into, or "differentiate," into the various organs. How, exactly, does a cell in the gut tube know it should become part of our lung instead of part of our stomach?
UVA's Chongzhi Zang, Ph.D., and collaborators have found answers, revealing how genetic material called chromatin interacts with other factors to switch genes on and off to carry out this essential transformation.
"Gut development is a fascinating dynamic process, from which we can learn how the same genome can create many different types of cells in different organs," said Zang, of UVA's Center for Public Health Genomics and UVA Cancer Center. "We knew the genes being used in different organs would start to show some differences in the early stages of development, but this was the first time that we found how such differences were controlled by chromatin during the organ-formation process."
Understanding organ development
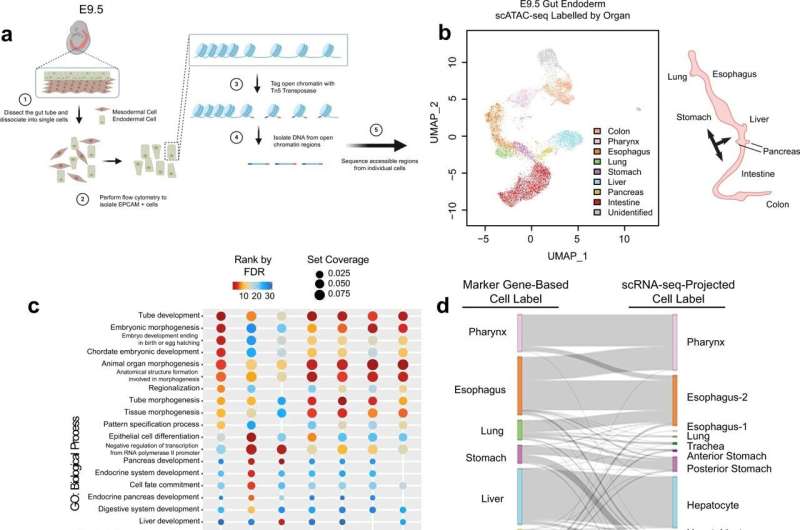
Zang and his colleagues, including collaborators led by Tae-Hee Kim, Ph.D., of the University of Toronto in Canada, used cutting-edge genomics technology called "single-cell ATAC-seq" to create a detailed "map" of the chromatin pattern changes that take place inside individual cells in the gut tube during organ formation in mice. In so doing, they have filled in many important gaps in our understanding of the organ-development process in mammals.
The team found that chromatin displays different dynamics in cells that become the liver, for example, than in cells that become the lungs. Chromatin interacts with what are called "transcription factors" in an elegant arrangement that trains the cells for the important jobs they are fated to hold.
Later in development, these interactions will further refine the emerging organs, allowing the intestine, for example, to subdivide into the large intestine and the small intestine.
It's important that this complex process play out precisely. The researchers found that errors can have dire consequences, disrupting, for example, the healthy development of the pancreas and intestines in lab mice. Dramatic changes noted in the pancreas included the formation of many large, cyst-like structures.
The researchers note that "cell fate" errors occur in the early stages of pancreatic cancer, leading to precancerous lesions. So understanding the organ-development process and what can go wrong could offer important insights into the formation of certain cancerous tumors.
"A better understanding of how genes work in the genome during organ development can give us insights into the mechanisms underlying initiation of many types of cancer," Zang said. "We use state-of-the-art technologies to tackle these complex problems and believe that these fundamental discoveries, one step at a time, will eventually inspire new therapeutic development and benefit cancer patients in the future."
The researchers have published their findings in Nature Communications.
More information: Ryan J. Smith et al, Single-cell chromatin profiling of the primitive gut tube reveals regulatory dynamics underlying lineage fate decisions, Nature Communications (2022). DOI: 10.1038/s41467-022-30624-w
Journal information: Nature Communications
Provided by University of Virginia