Study compares two methods for distance measurement in motile proteins

In the Middle Ages, every city had its own system of measurement. Even today, you can sometimes find iron rods in marketplaces that determined the length measurement valid for the city at that time. In science, however, there is no room for such uncertainties, and no matter what method you use to measure the length of a molecule, for example, the answer should always be the same. Researchers at the University Hospital Bonn (UKB), the University of Bonn and Ludwig-Maximilians-Universität (LMU) Munich have now investigated whether this is true for two methods that are very often used to measure distances in protein molecules—for example, to find out how such molecules move. The study has now appeared in the journal Nature Communications.
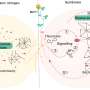
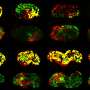
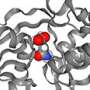
The researchers from PD Dr. Gregor Hagelueken's group at the Institute of Structural Biology of the UKB used so-called PELDOR spectroscopy to study the movement of so-called substrate-binding proteins. These proteins grab their substrate and transport it to a specific location in the cell. To observe this precisely, the researchers attached tiny magnets—the researchers call them "spin labels"—to the proteins and measured distances that are only about a billionth of a meter long. They then transmitted their results to the research group of Prof. Dr. Thorben Cordes at LMU Munich. There, comparative measurements were carried out using so-called FRET spectroscopy. Tiny dye molecules were used instead of spin labels.
"Although both methods are used very frequently, no one has yet systematically investigated whether the results are really comparable," says Hagelueken. Although it turned out that the measurement results were comparable in most cases, the researchers encountered inconsistencies in two cases. Bonn post-doctoral researcher Martin Peter says, "We then thoroughly investigated what caused the differences and found what we were looking for. In one case, it turned out that the dye molecules stuck to the protein and thus falsified the measurement." In the second case, the addition of a type of antifreeze, which was necessary because of the low measurement temperature of below -220 degrees Celsius, led to unexpected deviations. "We were able to show that despite the high accuracy of the methods, re-measuring with another nano ruler is always a good idea," Hagelueken says.
More information: Martin F. Peter et al, Cross-validation of distance measurements in proteins by PELDOR/DEER and single-molecule FRET, Nature Communications (2022). DOI: 10.1038/s41467-022-31945-6
Journal information: Nature Communications
Provided by University of Bonn