Inspiration and thinking in the design of large-scale functional proteins: Evolution-guided atomistic design
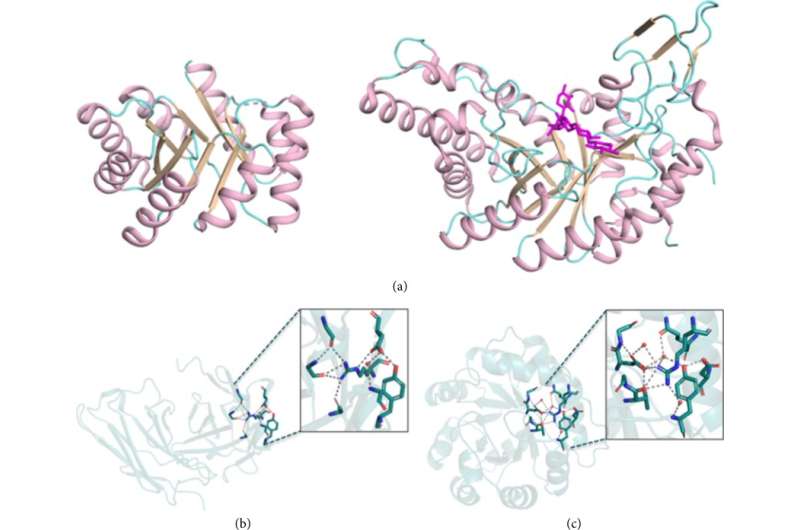
On 08 March 2022, the journal BioDesign Research published a review article titled "What Have We Learned from Design of Function in Large Proteins?" by Olga Khersonsky and Sarel J. Fleishman. The article proposes an evolution-guided atomistic design approach that eliminates mutations that could destabilize proteins, induce misfolding, or distort the active site domain of residues, and preserves non-ideal key sequences and structural features through the use of natural backbone and sequence constraints.
The authors first summarize key points in the design of large proteins. One fundamental reason why proteins are large is that many sophisticated binders and enzymes must encode unstable molecular features in their active sites, and stabilizing these features requires significant thermodynamic compensation from large regions outside the active site. The authors developed a general strategy for designing protein backbones in large proteins by assembling subdomain fragments. In this method, after backbone fragment assembly, atomic sequence design of the entire protein is performed. During this stage, the authors do not allow the selection of all amino acids at every position, but instead bias the design calculations towards mutations that are common among homologues, prohibiting rare mutations.
The authors then proposed a reliable and fully computational protein optimization approach and developed two different strategies: PROSS was used to optimize the stability of the protein in its native state, and FuncLib was used to design the amino acid residues of the active site or protein binding sites. To verify the usability of these strategies, the authors tackled a "hard problem" that was difficult to solve with iterative methods by testing proteins that could only be measured at low throughput.
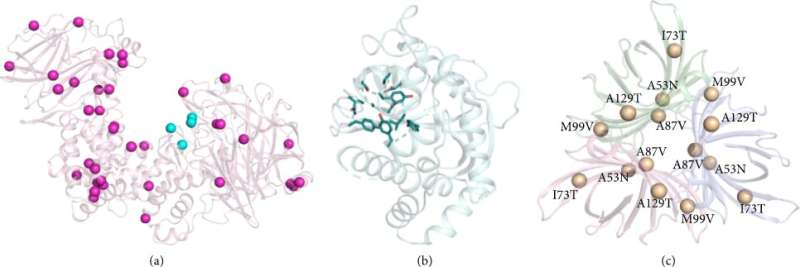
Finally, by reviewing the method development process for evolution-guided atomistic design, the authors explained why this approach is reliable and described the new design principles that have been revealed through its application. "Thus, the next phase of innovation in protein design methodology is likely to rely in part on statistical learning methods. These may open the way to one of the most long-standing goals of protein engineering: the completely computational design of new or improved molecular activities without recourse to experimental data," the authors wrote.
More information: Olga Khersonsky et al, What Have We Learned from Design of Function in Large Proteins?, BioDesign Research (2022). DOI: 10.34133/2022/9787581
Provided by Nanjing Agricultural University The Academy of Science