Self-organization of complex structures
Ludwig Maximilian University of Munich researchers have developed a new strategy for manufacturing nanoscale structures in a time- and resource-efficient manner.
Macromolecules such as cellular structures or virus capsids can emerge from small building blocks without external control to form complex spatial structures. This self-organization is a central feature of biological systems. But such self-organized processes are also becoming increasingly important for the building of complex nanoparticles in nanotechnological applications. In DNA origami, for instance, larger structures are created out of individual bases.
But how can these reactions be optimized? This is the question that LMU physicist Prof. Erwin Frey and his team are investigating. The researchers have now developed an approach based on the concept of time complexity, which allows new strategies to be created for the more efficient synthesizing of complex structures, as they report in the journal PNAS.
A concept from the computer sciences
Time complexity originally describes problems from the field of informatics. It involves investigating how the amount of time needed by an algorithm increases when there is more data to process. When the volume of data doubles, for example, the time required could double, quadruple, or increase to an even higher power. In the worst case, the running time of the algorithm increases so much that a result can no longer be output within a reasonable timeframe.
"We applied this concept to self-organization," explains Frey. "Our approach was: How does the time required to build large structures change when the number of individual building blocks increases?" If we assume—analogously to the case in computing—that the requisite period of time increases by a very high power as the number of components increases, this would practically render syntheses of large structures impossible. "As such, people want to develop methods in which the time depends as little as possible on the number of components," explains Frey.
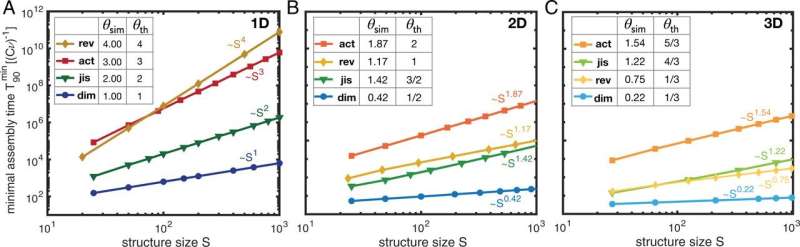
The LMU researchers have now carried out such time complexity analyses using computer simulations and mathematical analysis and developed a new method for manufacturing complex structures. Their theory shows that different strategies for building complex molecules have completely different time complexities—and thus also different efficiencies. Some methods are more, and others less, suitable for synthesizing complex structures in nanotechnology. "Our time complexity analysis leads to a simple but informative description of self-assembly processes in order to precisely predict how the parameters of a system must be controlled to achieve optimum efficiency," explains Florian Gartner, a member of Frey's group and lead author of the paper.
The team demonstrated the practicability of the new approach using a well-known example from the field of nanotechnology: The scientists analyzed how to efficiently manufacture a highly symmetrical viral envelope. Computer simulations showed that two different assembly protocols led to high yields in a short window of time.
A new strategy for self-organization
When carrying out such experiments before now, scientists have relied on an experimentally complicated method that involves modifying the bond strengths between individual building blocks. "By contrast, our model is based exclusively on controlling the availability of the individual building blocks, thus offering a simpler and more effective option for regulating artificial self-organization processes," explains Gartner. With regard to its time efficiency, the new technique is comparable, and in some cases better, than established methods. "Most of all, this schema promises to be more versatile and practical than conventional assembly strategies," says the physicist.
"Our work presents a new conceptual approach to self-organization, which we are convinced will be of great interest for physics, chemistry, and biology," says Frey. "In addition, it puts forward concrete practical suggestions for new experimental protocols in nanotechnology and synthetic and molecular biology."
More information: Florian M. Gartner et al, The time complexity of self-assembly, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2116373119
Journal information: Proceedings of the National Academy of Sciences
Provided by Ludwig Maximilian University of Munich