Researchers unlock behavior of rare cells that evade antibiotics by 'playing dead'
Researchers have developed a new microfluidic platform to track a very rare type of cell that can survive antibiotic treatments. The results have important implications for microbiologists learning about the cellular control of bacterial physiology and for scientists looking to combat the emerging threat of antibiotic resistance bacteria that can lead to untreatable infections.
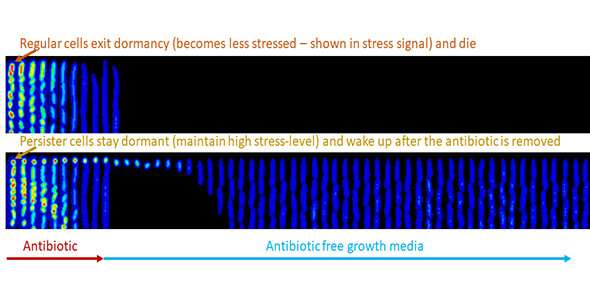
"Persister cells" are a very smallsubpopulation of cells that survive antibiotic treatment by staying dormant and then resume growth after the antibiotic has been removed. These cells are transient and are therefore harder to study and have no genetic changes, meaning that they cannot be identified using sequencing.
Since these "persister cells" are both rare and transient, a platform is needed to image cells over time with sufficiently large throughput. Now researchers from the University of Cambridge, Harvard Medical School and Harvard University have developed a platform designed to track more than 100,000 individual bacterial cells over hundreds of generations as they experience rounds of feast-and-famine cycles. An array of hundreds of thousands of traps are used to keep individual cells immobilized and infused with the contents from a growing culture—E. coli (Escherichia coli), normally found in the intestine, and Bacillus subtilis, commonly found in soil. A high-resolution microscope rapidly scans over each cell and tracks their fate over time. The results are reported in the journal Nature Microbiology.
Our microfluidic platform enables us to track cell lineages along the growth curve with precision and control, allowing us to learn more about how bacteria thrive and survive in complex changing environments," said co-author Dr. Somenath Bakshi, Lecturer in Synthetic Biology at the Department of Engineering, University of Cambridge. "We believe that it is this ability to track the growth, death and expression patterns of individual cells over time in dense, complex and changing cultures—potentially containing entire ecosystems—that could be transformative in many areas of microbiology.
"We can now study how the cells change their size and content to not only adapt to their current circumstances, but also to prepare for the unforeseen future. Furthermore, our research shows how these cells decide not to 'wake up' upon food replenishment and instead stay dormant to persist through antibiotic treatment.
"These experiments reveal a fascinating kaleidoscope of physiological palettes of cells under changing environments and give clues to how the diversity of fate outcomes of individual cells can have important benefits for the fate of the entire cell population. Studying 'persister cells'—a reservoir for the emergence of antibiotic-resistant cells—can be a useful first step in developing treatment strategies."
More information: Somenath Bakshi et al, Tracking bacterial lineages in complex and dynamic environments with applications for growth control and persistence, Nature Microbiology (2021). DOI: 10.1038/s41564-021-00900-4
Journal information: Nature Microbiology
Provided by University of Cambridge





















