Unraveling DNA packaging: How histones and DNA interact
The genetic material of most organisms is carried by DNA, a complex organic molecule. DNA is very long—for humans, the molecule is estimated to be about 2 m in length. In cells, DNA occurs in a densely packed form, with strands of the molecule coiled up in a complicated but efficient space-filling way. A key role in DNA's compactification is played by histones, structural-support proteins around which a part of a DNA molecule can wrap. The DNA-histone wrapping process is reversible—the two molecules can unwrap and rewrap—but little is known about the mechanisms at play. Now, by applying high-speed atomic-force microscopy (HS-AFM), Richard Wong and colleagues from Kanazawa University (NanoLSI WPI) provide valuable insights into the spatiotemporal dynamics of DNA-histone interactions.
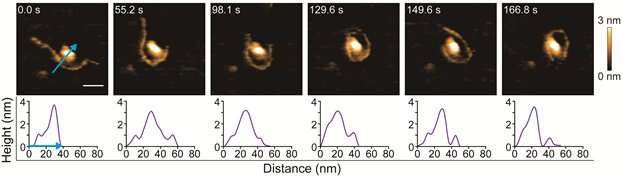
The researchers looked at the interaction between DNA and a histone called H2A, one of the five main histones. To check the applicability of HS-AFM as a viable tool for imaging the DNA-histone interaction, they first focused on H2A in its native state. Wong and colleagues were able to image the topology of the molecule, and how it changes over time. Importantly, they showed that the HS-AFM process, during which a tapping force is constantly exerted on the molecule, does not lead to conformational changes or actual damage.
For real-time observation of the DNA-H2A interaction with HS-AFM, the scientists prepared DNA samples with different lengths and forms: plasmid (long and circular), long-linearized and short-linearized DNA, with the latter having the highest motility. The experiments showed that the choice of substrate on which to put the DNA for AFM imaging is crucial; a particular type of lipid layer was found to be good as it does not strongly absorb DNA strands.
The observations of the interaction of H2A with short-linearized DNA, which the researchers nicknamed 'inchworm DNA', led to the most notable results. Specifically, four different interaction situations could be distinguished: touching, sliding, sandwiching and wrapping, with the associated motions indeed resembling the movements of inchworms.
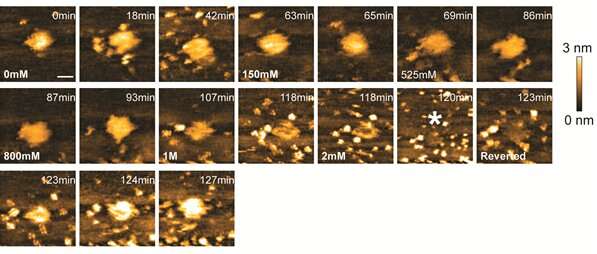
Wong and colleagues also investigated the effect of ionic strength on the DNA-histone binding affinity, by changing the salt concentration of the liquid containing the DNA-histone aggregate. When increasing the liquid's salinity, the aggregate was found to dissolve. When diluting the liquid again—and so reducing the salt content—the aggregate reformed. This result shows that varying the ionic strength (i.e., the salt concentration) of the environment of the DNA-H2A complex provides a way to mimic the variations in the strength of DNA-histone interactions as they happen in living organisms.
The report of Wong and colleagues represents the first real-time observation of DNA-histone interactions, and convincingly shows the applicability of HS-AFM for studying this kind of biological process, also in the context of diseases. Quoting the researchers: "[Our work] demonstrates ... the potential to study protein aggregation and protein-nucleic acid aggregate formation in various human diseases."
Finally, it is worth highlighting the contribution of the paper's first author, Goro Nishide, who is a pre-doctoral student at the Division of Nano Life Science in the Graduate School of Frontier Science Initiative at Kanazawa University. Mr Nishide played a key role in the reported research, supervised by Professor Wong and Dr. Lim, by performing the experiments, co-designing the study and co-writing the paper. Mr Nishide is also enrolled in Kanazawa University's WISE program for Nano-Precision Medicine, Science and Technology, an initiative aimed at innovations in disease prevention, diagnosis, and treatment methods based on exploiting our increased understanding of biological and other processes at the nanoscale.
More information: Goro Nishide et al, High-Speed Atomic Force Microscopy Reveals Spatiotemporal Dynamics of Histone Protein H2A Involution by DNA Inchworming, The Journal of Physical Chemistry Letters (2021). DOI: 10.1021/acs.jpclett.1c00697
Journal information: Journal of Physical Chemistry Letters
Provided by Kanazawa University