At the crossroads of cell survival and death
National University of Singapore researchers discovered that a protein, known as MOAP-1, plays a crucial role in facilitating autophagy, a cellular "self-eating" process that recycles non-essential components during starvation.
Autophagy, coined from the Greek words "auto" and "phagein" which mean "self" and "to eat," respectively, is an evolutionarily conserved cellular process that was originally identified as a survival mechanism used by yeast to survive under a harsh starvation condition by breaking down intracellular components that serve as an alternate source of energy. Mounting evidence, however, now suggests that autophagy also plays pivotal roles in maintaining cells in a healthy state against internal and external stresses such as carcinogenic insults and metabolic imbalance. Improper functioning of autophagy has been implicated in accelerated aging, and perhaps not surprisingly, also linked to the accelerated development of a wide range of major diseases commonly associated with elderly populations, including cancers and neurodegenerative diseases. Many cellular components that are required for the execution of autophagy have already been identified through the work of many researchers. This includes Prof Yoshinori Ohsumi, who was awarded the Nobel Prize in Medicine and Physiology in 2016 for his groundbreaking work in yeast which opened up the field of research in autophagy. However, how the precision with which this process is regulated in humans is still unclear.
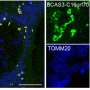
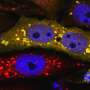
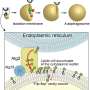
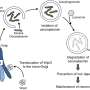
A team led by Prof Victor Yu at the Department of Pharmacy, NUS has found that a protein known as MOAP-1 is needed for the late step in the autophagy process, which involves the closure of the double membrane vesicle, known as autophagosome (see Figure). The intact autophagosome will fuse with the cellular degradative machinery, known as the lysosome, for degradation.
Through in-depth analysis, the research team found that the MOAP-1 protein physically interacts with the LC3 protein, which is a critical component of the membrane forming the autophagosome. Interestingly, interaction of LC3 with MOAP-1 is required for facilitating the final closure of the autophagosome. Hence, it is possible to stimulate autophagic activity through elevating levels of MOAP-1 protein to remove unwanted, damaged and potentially toxic cellular components such as protein aggregate deposits that cause neuron death in many neurodegenerative diseases.
Embarking on this exciting direction, the team will investigate the role of MOAP-1 in regulating selective forms of autophagy, including mitophagy and aggrephagy that concern the removal of damaged mitochondria and protein aggregates, respectively. Abnormal function in these processes have already been associated with accelerated aging and certain types of neurodegenerative diseases.
Prof Yu said, "It is fascinating to find that MOAP-1, which we previously demonstrated to be a protein promoting cell death, is also involved in facilitating autophagy to promote cell survival during starvation. The role of MOAP-1 in autophagy can certainly be further explored to promote understanding of the development of many aging-associated diseases such as cancers and dementia."
More information: Hao-Chun Chang et al. The BAX-binding protein MOAP1 associates with LC3 and promotes closure of the phagophore, Autophagy (2021). DOI: 10.1080/15548627.2021.1896157
Provided by National University of Singapore