New insights into evolution of gene expression
Vertebrate organs organize physiological activities, and the diverse expression patterns of thousands of genes determines organ identities and functions. Because of this, the evolution of gene expression patterns plays a central role in organismal evolution.
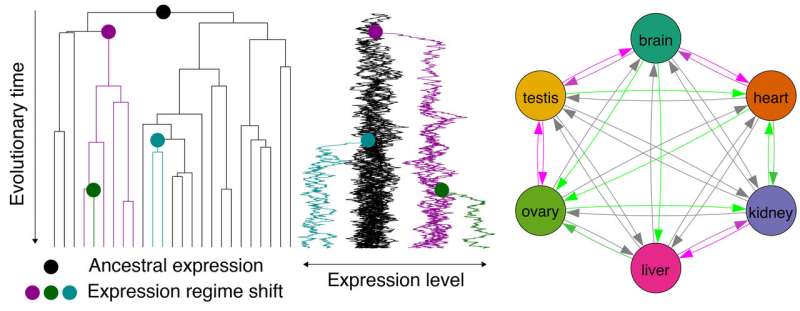
Major organ-altering evolutionary events such as development of the hominoid brain are also associated with gene expression shifts. However, although gene duplication is well-known to play an important role in expression pattern shifts, the evolutionary dynamics of expression patterns with and without gene duplication remain poorly understood.
"An important question is whether long-term expression in one organ predisposes genes to be subsequently utilized in other organs. The answer is yes," says Dr. Kenji Fukushima from Julius-Maximilians-Universität (JMU) Würzburg in Bavaria, Germany. "There are preadaptive propensities in the evolution of vertebrate gene expression, and the propensity varies with the presence and type of gene duplication."
Kenji Fukushima now reports this and other findings with his co-author David D. Pollock (University of Colorado School of Medicine, Aurora, U.S.) in the journal Nature Communications.
Dr. Fukushima holds a research position at the JMU Chair for Molecular Plant Physiology and Biophysics. Here, since 2018, the Japanese evolutionary biologist has been building up a working group, funded with 1.6 million euros by the Alexander von Humboldt Foundation. The renowned foundation had selected him as the winner of its Sofja Kovalevskaja Prize 2018. The award is intended for exceptionally talented young researchers.
Complex history of gene family trees
For their study the scientists amalgamated 1,903 RNA-seq datasets from 182 research projects. The date include six organs (brain, heart, kidney, liver, ovary, and testis) from 21 vertebrate species, ranging from freshwater fish and frogs to lizards, birds, rodents and humans. So they revealed a complex history of gene family trees. This allowed them to analyze the evolutionary expression of a broad set of genes.
More information: Nature Communications (2020). DOI: 10.1038/s41467-020-18090-8 , www.nature.com/articles/s41467-020-18090-8
Provided by University of Würzburg