Dock and harbor: A novel mechanism for controlling genes
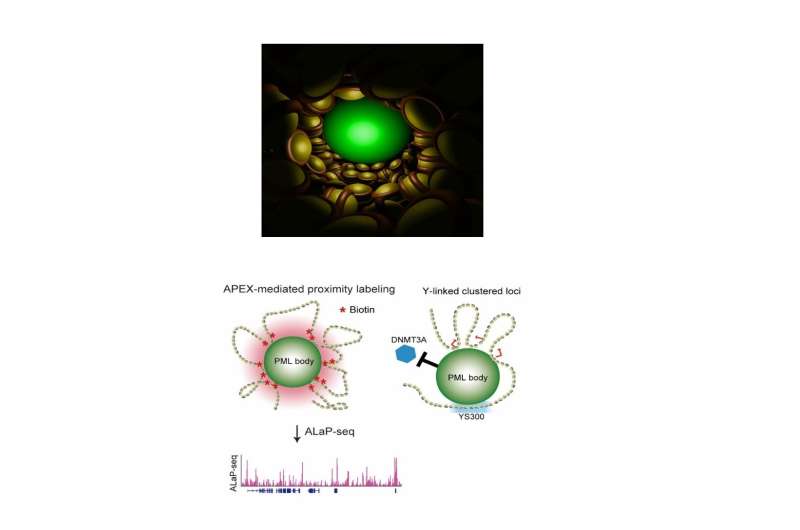
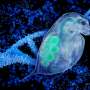
The genetic information within cells makes individuals unique. The cell nucleus has a complex structure that harbors this genetic information. The main component of the nucleus is chromatin, an intercalated pool of genes and proteins. Promyelocytic leukemia (PML) bodies are structures found closely associated with chromatin, suggesting that they may be involved with genetic function. However, the exact nature of the relationship between PML bodies and genes is unknown. In research conducted by a team at National Institutes of Natural Sciences (NINS) led by Yusuke Miyanari who recently joined NanoLSI at Kanazawa University, shows how PML bodies can modulate certain genes and the potential implications of these actions.
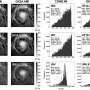
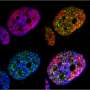
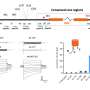
In order to visualize and track the exact location of PML bodies on the chromatin, the team developed a method known as APEX-mediated chromatin labeling and purification (ALaP). A fluorescent dye was first coupled with the PML bodies such that the light emitted by this dye could help trace the bodies. Subsequently, the PML body–chromatin complex could be isolated and the genes within it sequenced and identified.
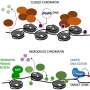
This technique was tested in cells of mice and resulted in successful extraction of the complex without any structural damage. The chromatin region anchored to the PML bodies within this complex was then identified as YS300, a short area of the Y chromosome. What's more, a cluster of genes in the vicinity of YS300 was also found to be impacted—some genes were suppressed and some were activated. PML bodies were thus somehow controlling the activity of these neighboring genes, which prompted the team to try and understand how.
In order for genes to be activated they must undergo a process known as DNA methylation. However, the structures of the suppressed genes suggested that they had been deprived of this process. A closer examination of the entire complex revealed that PML bodies were docked onto YS300 in a manner which prevented DNMT3A, a core regulator of DNA methylation, from accessing the genes. PML bodies were therefore physically restricting DNMT3A thereby preventing genetic activation.
"Our study sheds light on a newly discovered role of PML bodies in the regulation of the cluster genes and revealed a novel mechanism to regulate gene expression by 3-D nuclear organization," summarize the researchers. PML bodies are heavily involved in nervous system development, stress responses, and cancer suppression. Their newly found role can help understand whether and how gene regulation is involved in any of these cellular processes. Additionally, PML bodies can also be exploited as a potential switch to control the activity of certain genes.
More information: Misuzu Kurihara et al. Genomic Profiling by ALaP-Seq Reveals Transcriptional Regulation by PML Bodies through DNMT3A Exclusion, Molecular Cell (2020). DOI: 10.1016/j.molcel.2020.04.004
Journal information: Molecular Cell
Provided by Kanazawa University