Study shows evolution turns genes back on to regain function
Genes often mutate and lose their natural or synthetic function over long-term evolution, which could be good if that stops drug resistance of infectious microbes or cancer. A new study by Stony Brook University researchers, published online in PNAS, shows that evolution can exploit positive feedback (PF) within cells to restore gene function. Such repair by evolution may provide a basis for regaining lost gene function, which has implications in medicine and other scientific endeavors.
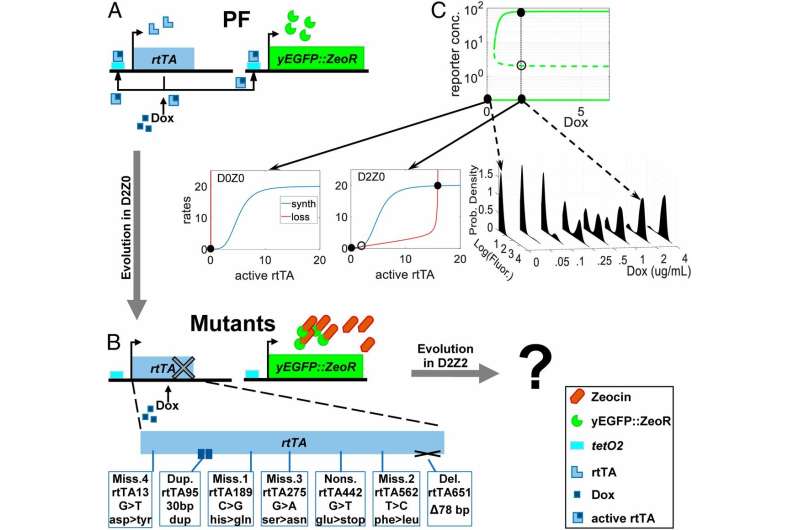
Based on the idea and experiments of an undergraduate Biomedical Engineering student, Mirna Kheir, and led by Gábor Balázsi, Ph.D., the Henry Laufer Associate Professor in Stony Brook University's Laufer Center for Physical and Quantitative Biology, and Department of Biomedical Engineering, the study included using synthetic PF in yeast cells by way of a chromosomally integrated gene circuit to test the process of regaining lost gene functions.
"We showed through these experiments and computational models that many drugs can activate mutant resistance genes through this process," explains Balázsi. "Essentially we exposed mutant, drug-sensitive cell populations to conditions where regaining resistance would be beneficial, and we found adaptation scenarios with or without repairing lost gene circuit function."
The results also suggest that inactive, nonfunctional natural drug resistance modules can also regain function upon drug treatment, quickly converting drug-sensitive cancer cells or microbes in drug-resistant ones.
More information: Mirna Kheir Gouda et al. Evolutionary regain of lost gene circuit function, Proceedings of the National Academy of Sciences (2019). DOI: 10.1073/pnas.1912257116
Journal information: Proceedings of the National Academy of Sciences
Provided by Stony Brook University