Tracking down the functions of the microbiome

All living creatures—from the simplest animal and plant organisms right up to the human body—are colonized by numerous microorganisms. They are thus in a functional relationship with these microbes, and together form a so-called metaorganism. The investigation of this symbiotic cooperation between host organism and microorganisms is a key challenge for modern life sciences research. The composition of the microbiome, i.e. the totality of the microorganisms which colonize a body, is well studied in numerous organisms.
However, how these microbes cooperate with the host, and what role they play in its biological functions, is still largely unknown. The Collaborative Research Centre (CRC) 1182 "Origin and Function of Metaorganisms" at Kiel University (CAU) aims to understand the communication and thus also the functional consequences of host-microbe interactions. Researchers at the CRC 1182 have now, for the first time, presented a functional repertoire of the microbiome of the nematode (or thread worm) Caenorhabditis elegans, thereby providing new insights into the connection between bacterial properties and metabolic functions of the host. The Kiel scientists recently published their results, which could also serve as a model for the analysis of the microbiome functions in other organisms, in the ISME Journal.
Nematode microbiome provides all important nutrients
Due to its simple structure and short generation time, the nematode C. elegans is a classic model organism in biological and medical research. However, to date, the role of its microbial symbionts has been mostly overlooked. Only a few years ago, a research team from the Evolutionary Ecology and Genetics research group at CAU characterized, for the first time, the natural bacterial colonizers of this worm. The Kiel research team also obtained first evidence of a functional influence of microorganisms, for example on the stress resistance of the worm or its ability to resist pathogens.
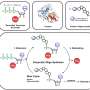
"On this basis, we wanted to gain an overview of the functional repertoire of the natural worm microbiome," said Nancy Obeng, doctoral researcher in the Evolutionary Ecology and Genetics research group and CRC 1182 member. "To do so, we predicted the metabolic networks of the nematode microbiome, based on whole genome sequence data from the bacteria," continued Obeng.

For this purpose, the CRC 1182 researchers analyzed the genome sequences of a total of 77 representative bacterial species from the digestive tract of the worm, and used these to infer metabolic networks of these microorganisms using mathematical modeling. This approach allowed them to predict which metabolites can arise as end products of certain available nutrients. "The conclusion of our modeling is that the microbiome of the worm is capable of producing nearly all essential nutrients for the host," said Johannes Zimmermann, doctoral researcher in the Medical Systems Biology research group at the CAU and also a member of the CRC. These results were subsequently confirmed experimentally in the laboratory. "It is primarily the frequently-occurring microbial colonizers, which provide the nematode with its required nutrient components," added Obeng.

Model for functional microbiome research
Using the example of the nematode C. elegans, the CRC 1182 researchers have thus developed a model to theoretically and experimentally derive metabolic networks on the basis of whole genome sequencing data. Similar to how the nematode serves as a model system for various life processes, such as individual development or the aging process, the methodology developed here could in future also help to determine the functional scope of the microbiome in other organisms—including humans.
"With our new approach for functional analysis of the microbiome, we have opened the door to a better understanding of the fundamental interaction of organisms and their microbial symbionts," said CAU Professor Hinrich Schulenburg, head of the Evolutionary Ecology and Genetics research group and Vice-Speaker of the CRC 1182. "In the future, we intend to perform comparable functional analyses across the entire spectrum of model organisms studied in our Collaborative Research Centre here in Kiel," added CAU professor Christoph Kaleta, head of the Medical Systems Biology group. Currently, the scientists from the Kiel metaorganism CRC are applying for renewed funding from the German Research Foundation (DFG), to continue exploring the interaction of organisms with their microbial partners.
More information: Johannes Zimmermann et al. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans, The ISME Journal (2019). DOI: 10.1038/s41396-019-0504-y
Journal information: ISME Journal
Provided by Kiel University