New genomics approaches to nomura's jellyfish sheds light on early evolution of active predation
A recent study, affiliated with South Korea's Ulsan National Institute of Science and Technology (UNIST) has analyzed the genetic map of Nomura's jellyfish, scientifically known as Nemopilema nomurai, for the first time.
The genetic map of Nomura's jellyfish, scientifically known as Nemopilema nomurai, has been analyzed for the first time. Through this, the researchers have provided clues to the genetic basis of the innovative structure, function, and chemistry that have allowed this distinctive early group of predators to colonize the waters of the globe.
Through this work, Professor Jong Bhak, founding director of Korean Genomics Industrialization Center (KOGIC), together with Korea Institute of Ocean Science and Technology (KIOST), have presented the completed genome sequence of the Nemopilema jellyfish. Analyses of the Nemopilema and other cnidarian genomes revealed adaptations associated with swimming, marked by codon bias in muscle contraction and expansion of neurotransmitter genes, along with expanded Myosin type II family and venom domains, possibly contributing to jellyfish mobility and active predation. They also identified gene family expansions of Wnt and posterior Hox genes and discovered the important role of retinoic acid signaling in this ancient lineage of metazoans, which together may be related to the unique jellyfish body plan (medusa formation).
Taken together, the Nemopilema jellyfish genome and transcriptomes genetically confirm their unique morphological and physiological traits, which may have contributed to the success of jellyfish as early multi-cellular predators.
Jellyfish are classified as a member of the phylum Cnidaria, a group that also includes hydra, sea anemones, and corals. Unique among cnidarians, jellyfish have remarkable morphological and biochemical innovations that allow them to actively hunt in the water column and were some of the first animals to become free-swimming.
"The cnidarians appear in an evolutionarily very important position, as it shares a common ancestor with the bilaterians, such as humans, fruit flies, and zebrafish," says Director Bhak of KOGIC. "As jellyfish are evolutionarily very distant among cnidarians, the genome of the giant Nomura's jellyfish sheds light on the early evolution of active predation."
The massive Nomura's jellyfish, whch can grow to two meters diameter with a weight of 200kg, is often found in the southern coast of South Korea. These creatures are toxic, thus can be a great threat to fisheries and ocean ecosystems. Since 2000, the giant Nomura's jellyfish has increased significantly, mainly due to global warming and a lack of natural predators. The Nemopilema nomurai genome has provided clues to the identification of toxin-related domains in jellyfish, as well as the prevention method for bulk breeding.
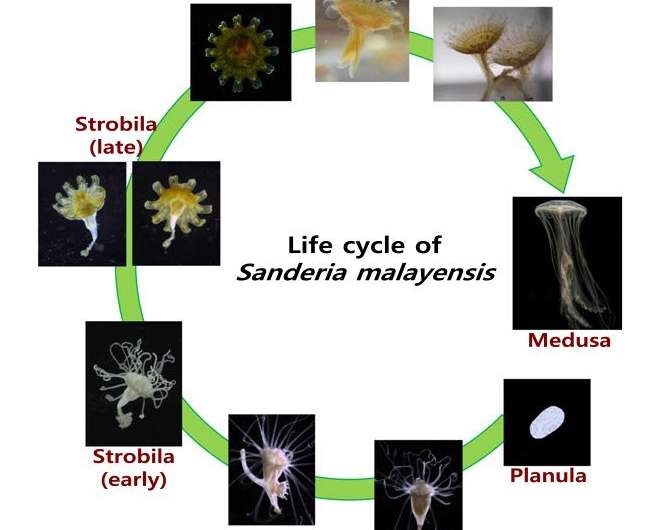
"In the case of jellyfish, a single adult polyp, called polyp, grows through transformation and growth processes to 5,000, so it is considered a fundamental measure to prevent large-scale reproduction of polyps," says Seungshic Yum in the Ecological Risk Research Division from Korea Institute of Ocean Science & Technology (KIOST). "Through this study, we were able to discover a gene related to signal transducer that plays an important role in polyp metamorphosis, and it could provide a basis for future prevention of mass reproduction of jellyfish."
Through this study, the researchers also obtained protein gene information for the nematocyst jellyfish poison. Jellyfish use tentacles to catch food and use poison at this time. The jellyfish genome analyzed here has identified a specific protein domain with increased numbers of poisons.
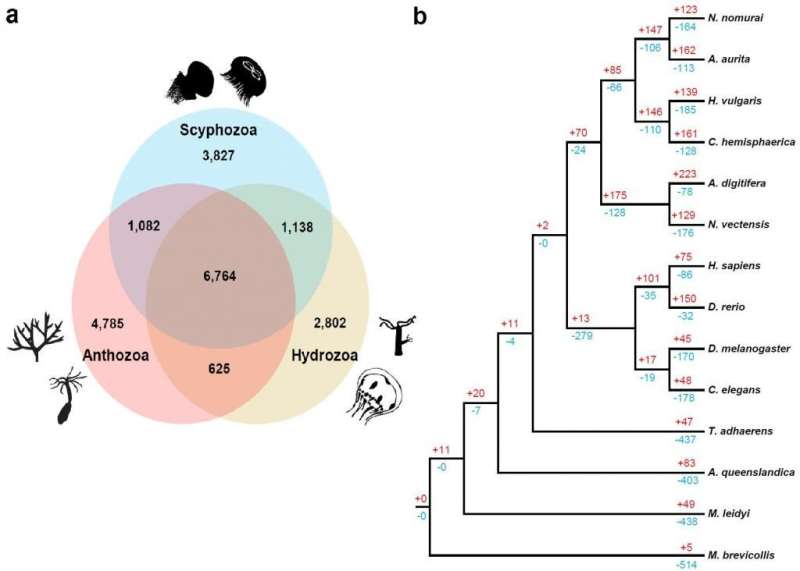
To identify jellyfish-specific evolutionary traits, the researchers examined gene family expansions and contractions across one unicellular holozoan and 13 metazoans, using 18,458 orthologous gene families. Among those, 10,434 were found in Nemopilema and 6,764 were shared by all three available cnidarian classes. Their findings suggest that jellyfish exhibits greater control over osmotic responses at the molecular level, when compared with other cnidarians. This can be interpreted that jellyfish have evolved to survive even when the total salt concentration in the ocean varies when they move vertically or horizontally to catch food and result in difference in total salt concentration in the ocean. Besides, the number of genes involved in muscle contraction was higher in jellyfish than in other cnidarians. The expression of these genes was higher in the medusa bell, which is essential for swimming.
"Some species of jellyfish are known to have infinite lifespans, which can also be helpful in the study of extreme nti-aging that could make reverses aging a reality," says Professor Bhak. "Such findings will be used in molecular biological studies for extreme anti-aging, along with the standard reference genome map for whales, tigers, and others that have been carried out at KOGIC."
More information: Hak-Min Kim et al. The genome of the giant Nomura's jellyfish sheds light on the early evolution of active predation, BMC Biology (2019). DOI: 10.1186/s12915-019-0643-7
Journal information: BMC Biology