Exploiting parasitic yeast to kill yeast pathogens
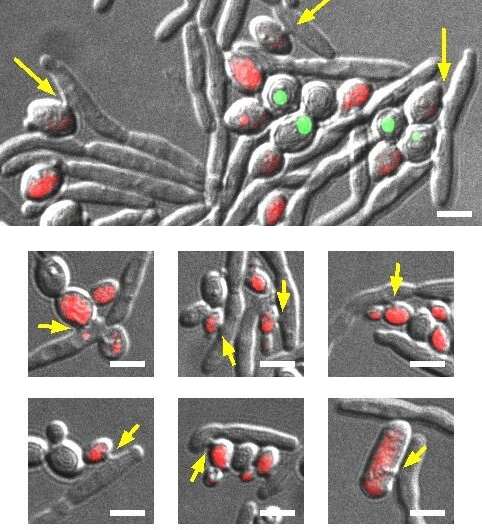
Insights into the genes and proteins involved in the predatory behavior of a parasitic yeast species could lead to new strategies for controlling yeast pathogens, according to a study published May 9 in the open-access journal PLOS Pathogens by Jürgen Wendland of the Carlsberg Research Laboratory in Denmark and Vrije Universiteit Brussel in Belgium, and colleagues.
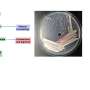
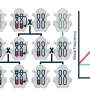
Pathogenic yeasts and fungi are an increasing global healthcare burden, but the discovery of novel antifungal agents is slow. The parasitic yeast Saccharomycopsis schoenii was recently demonstrated to be able to kill the emerging multi-drug resistant yeast pathogen Candida auris. However, the molecular mechanisms involved in the predatory activity of S. schoenii have not been explored. To this end, the researchers sequenced, assembled and annotated a draft genome of S. schoenii. They integrated quantitative live-cell microscopy assays with genomic, transcriptomic and proteomic approaches to identify genes and proteins that are overexpressed by S. schoenii during its predation of the model prey cell Saccharomyces cerevisiae.
The researchers hypothesized that the need for organic sulfur compounds, especially methionine, play a central role in the predatory behavior of S. schoenii. Surprisingly, they found that a general nutrition limitation, not a specific methionine deficiency, triggered predatory activity. During predation, both proteomic and transcriptomic analyses revealed that S. schoenii highly upregulated and translated genes that encode enzymes called aspartic proteases, probably used to break down prey cell walls. According to the authors, these fundamental insights into the predatory behavior of S. schoenii open up new avenues to exploit this yeast as a biocontrol agent or source for novel antifungal agents.
The authors add, "With the combined efforts of big-data global analysis tools we were able to place Saccharomycopsis schoenii into a CTG clade and dissect predacious behavior of this yeast to identify genes and pathways that likely play a decisive role in the predation process. Additionally, with 'caught-in-the-act' in vivo time lapse microscopy we were able to monitor predacious behavior and prey cell death over time."
More information: Junker K, Chailyan A, Hesselbart A, Forster J, Wendland J (2019) Multi-omics characterization of the necrotrophic mycoparasite Saccharomycopsis schoenii. PLoS Pathog 15(5): e1007692. doi.org/10.1371/journal.ppat.1007692
Journal information: PLoS Pathogens
Provided by Public Library of Science