November 21, 2018 feature
Cryogenic-electron microscopy (Cryo-EM) structures of a human ABCG2 mutant transporter protein
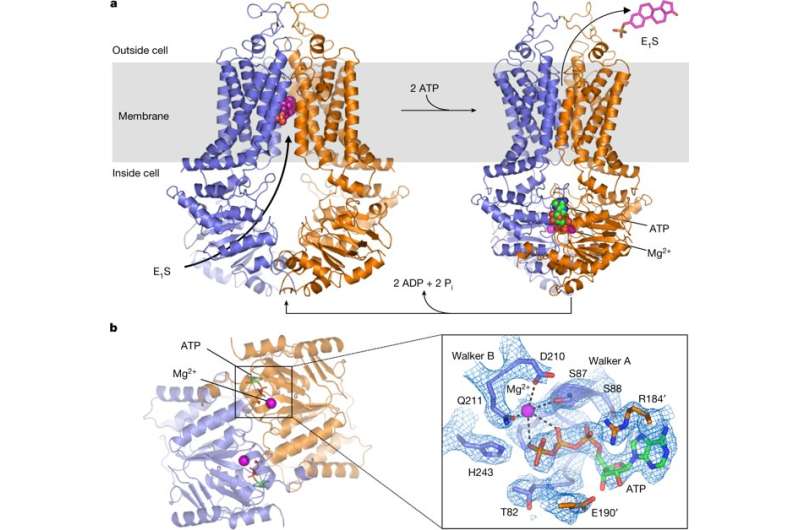
The transporter protein ABCG2 belongs to the ATP-binding-cassette (ABC) family. The protein is expressed in the plasma membranes of cells within a variety of tissues and tissue barriers, including the blood-brain, blood-testes and maternal-fetal barrier. The protein can be powered by ATP to translocate endogenous substrates, affect the pharmacokinetics of many drugs and protect against a variety of xenobiotics including anticancer drugs, notably in breast cancer. ABCG2 is often referred to as the breast cancer resistance protein, where previous studies have revealed the ABCG2 architecture and structural basis of ABCG2 inhibition with small molecules and antibodies. The mechanism of substrate recognition by ABCG2 alongside its ATP-driven transport capability remains yet to be determined.
In a new report now published in Nature, Ioannis Manolaridis and colleagues presented high-resolution cryoelectron microscopy structures of human ABCG2 in its substrate-bound state of pre-translocation and in the ATP-bound post-translocation state. The scientists used a mutant protein containing a glutamine in place of the catalytic glutamate (ABCG2EQ) to observe both states. The mutant ABCG2EQ showed reduced ATPase and transport rates to enable conformational trapping suited for the structural studies. In the substrate-bound state of the study, about halfway across the membrane in a cytoplasm-facing cavity, a single molecule of estrone-3-sulfate (E1S) was integrated for functional studies.
Rigid-body shifts of the transmembrane domain were observed as a result of ATP-induced conformational changes, pivoting the nucleotide-binding domains (NBDs) and changes in the relative NBD subdomain orientation. In its mechanism of action, ABCG2 harnessed the energy of ATP binding to extrude E1S and other endogenous substrates. Based on the size and binding affinity of compounds it was therefore possible to distinguish substrates from inhibitors in the study.
The authors first established that replacing the catalytic glutamate E211 with a glutamine in the Walker B motif (phosphate binding sequence) would result in mutants with greatly diminished (although not abolished), ATP hydrolysis and E1S transport activity. To determine the E1S bound structure, an antigen-binding fragment of monoclonal antibody 5D3 (5D3-Fab) was added to the sample – which bound to the external surface of ABCG2, to facilitate high-resolution structure determination. The ATP hydrolysis was slowed down and transport activity of liposome-reconstituted ABCG2 inhibited by the addition of 5D3-Fab, without impacting the interaction between ABCG2 and E1S substrate. The ABCG2EQ complex revealed an inward-open conformation with an overall resolution of 3.6 Å to clearly visualize the transmembrane domain and the substrate binding cavity.
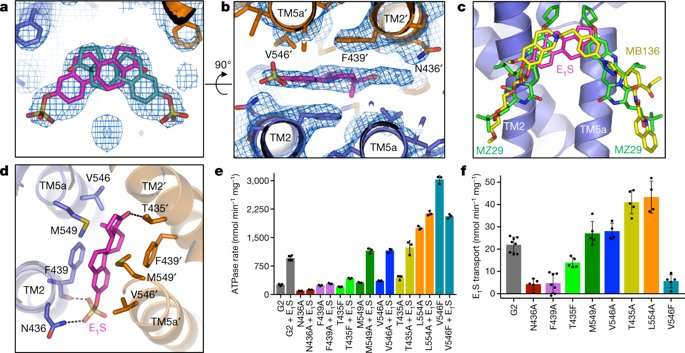
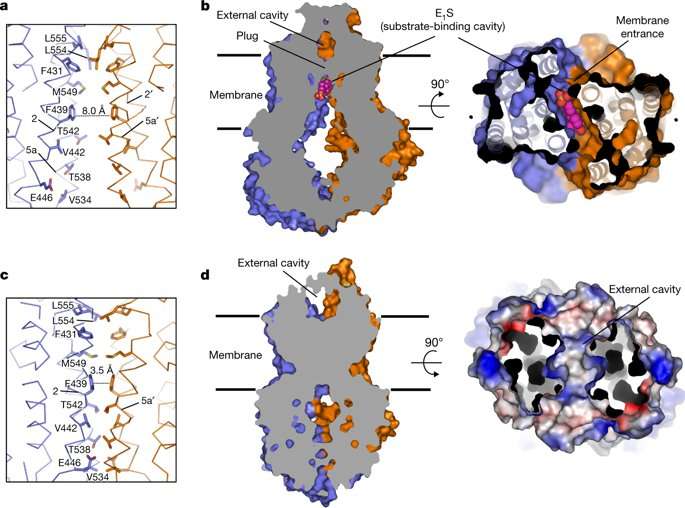
The authors observed a density feature formed by the transmembrane (TM) helices TM2 and TM5a in the substrate-binding cavity of ABCG2 monomers. The density feature could bind one E1S substrate in two orientations, although two E1S molecules could not be bound simultaneously due to steric clashing of the polycyclic ring systems. The substrate binding cavity could also accommodate inhibitors, demonstrating its dual role in substrate and multidrug binding.
In the study, the scientists generated single point mutations for ABCG2EQ- E1S to determine ATPase activity and E1S transport of the resulting mutant variants. The stability of all tested mutants showed similarity to the native, wild-type protein allowing direct comparison. The V546F mutant for instance, had impaired transport activity but a 12-fold increase in ATPase activity, although that was inhibited by E1S in a concentration-dependent manner. This result suggested that the introduction of two phenyl rings into the substrate cavity enabled substrate-binding mimicry for ATPase activity stimulation, but further inclusion of E1S 'clogged' the transporter. Results for other mutant variants varied in comparison to the wild-type protein in the study, emphasizing the sensitivity of the binding cavity to modifications.
Additional functional studies with cryo-EM microscopy showed that ATP binding caused a 35 degree rotation of the α-helical domains of NBD (nucleotide-binding domains) for dimerization—to enable the 'power stroke' in the transport cycle. In each ABCG2 monomer, individual transmembrane nucleotide binding domain interfaces remained largely unchanged between the nucleotide-free and ATP-bound states. However, ATP-induced conformational changes triggered shifts in NBD causing cytoplasmic components of the transmembrane domain (TMD) to push towards each other. Such ATP-induced conformational changes have important roles in the substrate-translocation pathway.
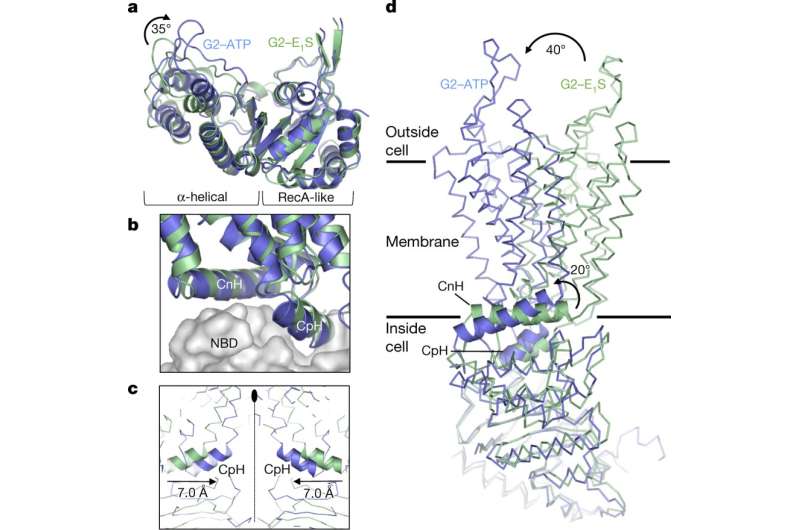
Structurally in the ATP-bound state of ABCG2EQ-EIS, phenyl rings stacked against each other, collapsing the substrate-bending cavity without space for the bound substrate. For transportation across the membrane, the substrates had to move through the translocation pathway at the center of the transporter to reach the external cavity before the pathway closed completely. For this process, transient conformational changes resembling peristaltic motion had to occur to generate space for the substrate.
The authors assumed that the substrate may bind via the cytoplasm or via the "membrane entrance" within the lipid bilayer. Once bound, the NBD dimer could only close when the substrate moved out of the substrate-binding cavity. In a productive transport cycle, the substrate was assumed to move through a translocation pathway at the center of the transporter via a "leucine plug." Once a substrate cleared the plug area, the structure of ATP-bound ABCG2EQ suggested that the plug region closed, and substrate was released to the outside.
One caveat in the study was that the E211Q mutation may have influenced the energetics of the conformational changes involved. The findings suggested that ATP binding may be sufficient for the substrate-extrusion step and the transporter could be reset to an inward-facing conformation by ATP hydrolysis. Unlike other transporters, the ABCG2 did not appear to form a stable, occluded conformation upon substrate binding, but demonstrated a transient conformation with peristalsis-like mechanisms akin to the bacterial BtuCD-F transporter.
A key question that had remained unanswered with polyspecific multidrug transporters such as ABCG2 was why some compounds acted as substrates, while others acted as potent inhibitors. When the authors compared the binding modes of ABCG2 substrate with those of two potent inhibitors, all three molecules bound to the same cavity of the transporter. The study further showed that when the substrate EIS and ATP bound together to ABCG2, an opening of the plug allowed the substrate to be pushed into the external cavity. In contrast, inhibitors acted as wedges to immobilize the transporters by locking the structure in an inward-facing conformation. The study suggested that the size and binding affinity of the compounds were important for translocation in the transporters, thereby allowing to distinguish substrates from inhibitors. The work provides new insight to further understand dynamics of the transporter protein ABCG2.
More information: Ioannis Manolaridis et al. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states, Nature (2018). DOI: 10.1038/s41586-018-0680-3
Scott M. Jackson et al. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2, Nature Structural & Molecular Biology (2018). DOI: 10.1038/s41594-018-0049-1
Zachary Lee Johnson et al. ATP Binding Enables Substrate Release from Multidrug Resistance Protein 1, Cell (2017). DOI: 10.1016/j.cell.2017.12.005
Journal information: Nature , Nature Structural & Molecular Biology , Cell
© 2018 Science X Network