Near-atomic resolution model of Ebola virus protein brings clearer understanding of the viral mechanics
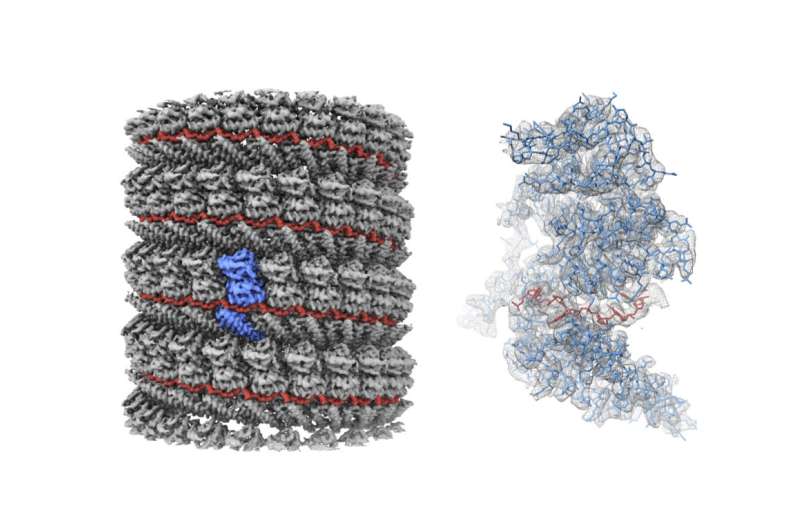
Researchers at the Okinawa Institute of Science and Technology Graduate University (OIST) have for the first time imaged the structure of a central component of the Ebola virus at near-atomic resolution.
The study, published in the journal Nature, was led by Prof. Matthias Wolf and primary author Dr. Yukihiko Sugita of OIST's Molecular Cryo-Electron Microscopy Unit (MCEMU) in a collaboration with world-renowned virologist Prof. Yoshihiro Kawaoka (University of Tokyo and University of Wisconsin-Madison), Prof. Takeshi Noda (Kyoto University) and structural biologist Dr. Hideyuki Matsunami (OIST).
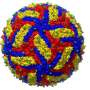
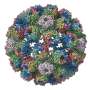
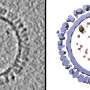
The researchers focused on a part of the virus called the nucleocapsid (NC), a complex of proteins that acts as a support structure for the viral genetic material. This structure allows the virus to replicate its deadly payload, to devastating effect. Dr. Sugita isolated nucleoprotein-RNA complexes, which make up the core of NCs, to a high degree of purity, then analyzed them with a cryo-electron microscope (cryo-EM) to take the closest-ever look at their structure.
"Before this study, we only knew about the smallest pieces of the NC structure," said Dr. Sugita, "Now that we can see it as a whole, it may help find targets for antiviral drugs."
The complete Ebola virus is a tiny invisible filamentous structure that appears like a short strand of hair when magnified thousands of times. Inside these filaments are the pieces that make up a functioning virus: NC and matrix proteins, both of which are wrapped by surface glycoproteins and a viral envelope.
But neither the scientists at OIST nor the public were in any danger from the study: the NP-RNA complex was produced without any actual viral RNA sequence in non-infectious cells grown in the biosafety-laboratory at OIST and did not represent any risk. "It's like the parts of a car," said Prof. Wolf, "Unless the chassis, engine, wheels and so on are all together, nothing works. That's what the particle we studied is like—most of its components are absent. It's harmless."
Previous studies had analyzed the NC structure using a technique called electron tomography, but the cryo-EM reconstruction generated by the OIST team has much higher definition, resolving individual RNA nucleotides and amino-acid side chains. "Our paper shows for the first time what the NC structure looks like at near-atomic resolution" said Prof. Wolf.
To get an idea of the step up the team made, imagine watching your favorite movie. If the previous study was like viewing the film on a VHS, then their novel structure is like seeing it in 4K presentation: the details are vastly sharper, with clear definition that allows scientists to get the most detailed picture yet of this component of the virus.
It was a slow process achieving this picture, however. "The protein complexes are very fragile," said Dr. Sugita, "getting a complete sample was very tricky."
Once the samples were prepared, it took the team more than 18 months to find just the right way to reconstruct the NC as a digital model. The helical structure of the protein complex is deceptively simple, but it has a fine complexity to it. Being off on the precise number and symmetry by just a few of the helix' tiny protein building blocks can invalidate the entire model. But after systematic search, a few hints provided by another team's publication, and new insight provided by their own high-resolution analysis, the OIST team's hard work paid off; everything slotted into place.
"Now that we have a clear picture of what this structure really looks like," said Prof. Wolf, "it is one step closer to figuring out how the whole virus works."
The three-dimensional model generated from the cryo-EM images provides an accurate foundation for future work. Researchers will now be able to make precisely targeted studies of the entire structure of Ebola virus NC, which may point to ways of defeating the virus.
More information: Yukihiko Sugita et al, Cryo-EM structure of the Ebola virus nucleoprotein–RNA complex at 3.6 Å resolution, Nature (2018). DOI: 10.1038/s41586-018-0630-0
Journal information: Nature
Provided by Okinawa Institute of Science and Technology