September 13, 2018 report
A way to detect likelihood of off-target cuts in CRISPR-Cas9
An international team of researchers has developed a way to detect the likelihood of making off-target cuts when using the CRISPR-Cas9 gene editing technique. In their paper published in the journal Nature, the group describes the new technique and how well it worked when tested.
CRISPR-Cas9 has held out promise of a revolution in gene editing for medical purposes, but has yet to live up to expectations. This is mostly because the technique has a serious flaw—it sometimes cuts non-targeted parts of DNA, which can, of course, be a very serious problem. In this new effort, the researchers have come up with a way of testing proposed guide RNA prior to use in CRISPR-Cas9 gene editing.
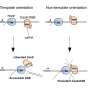
The idea behind this new research is to identify the parts of DNA that could be mistaken for a real target by guide RNA. If such parts are found, then a different guide RNA can be selected. This process can be repeated until a guide RNA is found that will only select the actual targets.
The testing method developed by the researchers consisted of cutting groups of base pairs from DNA strands from a test subject. Next, they applied adapters to circularize the DNA in each group. After that, they added a Cas9 nuclease and the proposed guide RNA—this step resulted in cuts to the DNA at certain sites. Then another pool of nucleases was used to degrade the circular DNA that did not get cut by the CRISPR-Cas-9, giving the researchers material to sequence for use in comparing places where cuts did occur. This allowed them to spot cuts that were not intended targets.
The researchers tested their method using mice. They found that their method detects guide RNA known to make off-target cuts. They also found that when they used guide RNA that had been found to produce no erroneous cuts, their test showed it was likely error-free.
The team sums up their work by suggesting the importance of selecting the right guide RNA for CRISPR-Cas9 editing to avoid off-target cuts—and their technique can ensure sure that happens.
More information: Pinar Akcakaya et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations, Nature (2018). DOI: 10.1038/s41586-018-0500-9
Journal information: Nature
© 2018 Medical Xpress