September 7, 2018 report
Two unrelated studies result in discovery of CRISPR-Cas12a inhibitors
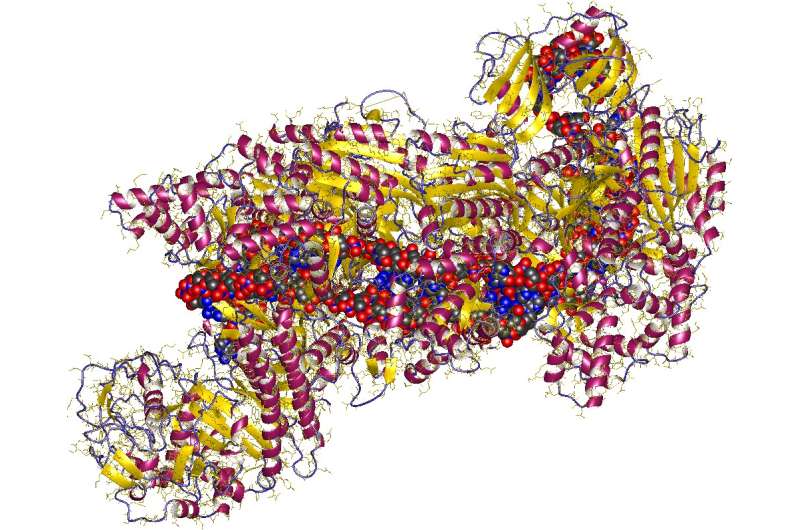
Two teams working independently of one another have identified several CRISPR-Cas12a inhibitors. The first team was made up of members from the University of California, Berkeley, the other had members from Massachusetts General Hospital and the University of California. Both used bioinformatics tools to scan bacterial genomes for possible inhibitors and both have published their results in the journal Science.
CRISPR is a gene editing technique that can identify DNA segments and snip them out of a genome. It can also be used to replace segments that have been cut out. In order to perform its duties, CRISPR uses a protein nuclease to serve as a guide or template, outlining which genes are to be cut and/or replaced. Cas9, often used for this purpose in many early studies, came to be associated with the technique. But for Cas9 to work properly, researchers also had to add in a Cas9 inhibitor—its job was to prevent extraneous cutting. More recently, researchers have used Cas12a because it offers other desired characteristics not found in Cas9. But until now, there were no known inhibitors for Cas12a, which stymied its use in research efforts. In these two new efforts, both teams have found several inhibitors that they claim are suitable for use with CRISPR.
Both teams used a bioinformatics pipeline approach in their search for Cas12 inhibitors—it is a system for searching through bacterial genomes that involves looking for genetic fragments that are normally deadly to bacteria but are not toward bacteria that have inhibitors. Using this approach, the first team found three inhibitors, including one that stood out called AcrVA1. Testing with human cells showed that all three could be used with CRISPR. Using the same basic approach, the second team found several candidates that also worked as hoped when tested with human cells. The second team also found AcrVA1 to be particularly effective.
Taken together, the work by the two teams has yielded several possible Cas12a inhibitors, one of which appears to be particularly promising. And that could lead to more fruitful research using CRISPR-Cas12a as a new gene editing tool.
More information: 1. Kyle E. Watters et al. Systematic discovery of natural CRISPR-Cas12a inhibitors, Science (2018). DOI: 10.1126/science.aau5138
Abstract
Cas12a (Cpf1) is a CRISPR-associated nuclease with broad utility for synthetic genome engineering, agricultural genomics, and biomedical applications. While bacteria harboring CRISPR-Cas9 or CRISPR-Cas3 adaptive immune systems sometimes acquire mobile genetic elements encoding anti-CRISPR proteins that inhibit Cas9, Cas3, or the DNA-binding Cascade complex, no such inhibitors have been found for CRISPR-Cas12a. Here we employ a comprehensive bioinformatic and experimental screening approach to identify three different inhibitors that block or diminish CRISPR-Cas12a-mediated genome editing in human cells. We also find a widespread connection between CRISPR self-targeting and inhibitor prevalence in prokaryotic genomes, suggesting a straightforward path to the discovery of many more anti-CRISPRs from the microbial world.
Nicole D. Marino et al. Discovery of widespread Type I and Type V CRISPR-Cas inhibitors, Science (2018). DOI: 10.1126/science.aau5174
Abstract
Bacterial CRISPR-Cas systems protect their host from bacteriophages and other mobile genetic elements. Mobile elements, in turn, encode various anti-CRISPR (Acr) proteins to inhibit the immune function of CRISPR-Cas. To date, Acr proteins have been discovered for type I (subtypes I-D, I-E, and I-F) and type II (II-A and II-C) but not other CRISPR systems. Here we report the discovery of 12 acr genes, including inhibitors of type V-A and I-C CRISPR systems. Notably, AcrVA1 inhibits a broad spectrum of Cas12a (Cpf1) orthologs including MbCas12a, Mb3Cas12a, AsCas12a, and LbCas12a when assayed in human cells. The acr genes reported here provide useful biotechnological tools and mark the discovery of acr loci in many bacteria and phages.
Journal information: Science
© 2018 Phys.org. All rights reserved.