Predicting the outcome of the arms race between man and bacteria
Through computer simulations, scientists can predict if bacteria can be stopped with popular antibacterial therapies or not – a breakthrough which will help select and develop effective treatments for bacterial infections.
Growing resistance to antibiotics is a one of the most serious global health threats we are facing. There is an urgent need to develop new antibiotics that are cost effective as it is estimated that by 2050, 10 million lives per year will be at risk from antibiotic-resistant infections. In a bid to help tackle this challenge, researchers at the University of Bristol have developed computer simulations which could be key to getting ahead in the ongoing 'arms race' with bacteria.
Researchers focused on enzymes in bacteria that can split the structure of penicillin-type antibiotics, leading to resistance. To restore the effectiveness of these antibiotics, 'resistance blocking' molecules have been developed to block the activity of these enzymes. By treating patients with the right combinations of antibiotics and resistance blockers, doctors are able to gain the upper hand in the battle. Unfortunately, bacteria can make many different enzymes able to destroy penicillins, and available resistance blockers work against only some of these.
New findings, published in Biochemistry, show that it is now possible to use computer simulations to predict whether these resistance blockers will be effective or not. It is hoped that this information will help scientists to develop improved resistance blockers, which can restore the action of popular antibiotics against a wider range of resistant bacteria.
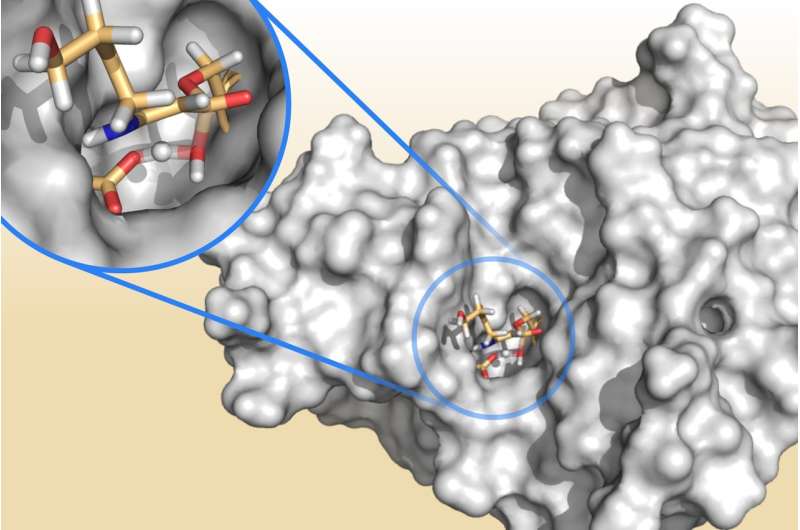
Using a computer simulation technique called QM/MM (quantum mechanics/molecular mechanics simulations) the Bristol research team were able to gain a molecular-level insight into how resistance enzymes react with resistance blockers.
Researchers specifically focused on clavulanic acid, a drug that prevents destruction of common penicillin-like antibiotics. Clavulanic acid is commonly used in combination with the antibiotic amoxicillin to treat ear, sinus and urinary tract infections (co-amoxiclav). Some such bacterial infections, however, are becoming much more difficult to treat because clavulanic acid does not work effectively against the enzymes that they produce.
The QM/MM simulations interrogated how clavulanic acid interacts with these bacterial enzymes and revealed the most important step in determining whether the enzyme is effectively blocked. An enzyme that cannot be stopped releases a broken-down molecule of clavulanic acid and goes on to break down the antibiotic with which it is administered, resulting in antibiotic resistance. If the breakdown of clavulanic acid takes a long time, then the enzyme gets 'clogged up' and is unable to break down the antibiotic, which can then kill the bacterium and clear the infection.
Dr. Marc van der Kamp, from Bristol University's School of Biochemistry, said:
"We are excited to see how our computer simulations can be used in the future to test enzymes from bacteria and predict when a resistance blocking inhibitor will be effective.
We hope that this will identify how we can better block such bacterial enzymes, so that antibiotics can be used effectively for treatment of drug resistant infections.
Our simulations may also be a valuable tool to help choose which combinations of drugs are best for treating a particular infection outbreak, allowing us to be better equipped in this 'arms race' with bacteria."
More information: Rubén A. Fritz et al. Multiscale Simulations of Clavulanate Inhibition Identify the Reactive Complex in Class A β-Lactamases and Predict the Efficiency of Inhibition, Biochemistry (2018). DOI: 10.1021/acs.biochem.8b00480
Journal information: Biochemistry
Provided by University of Bristol











.jpg)