Understanding a cell's 'doorbell'
A multi-institutional project to understand one of the major targets of human drug design has produced new insights into how structural communication works in a cell component called a G protein-coupled receptor (GPCRs), basically a "doorbell" structure that alerts the cell of important molecules nearby. Understanding the structure and function of the receptor more deeply will enable better drug development.
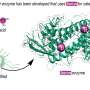
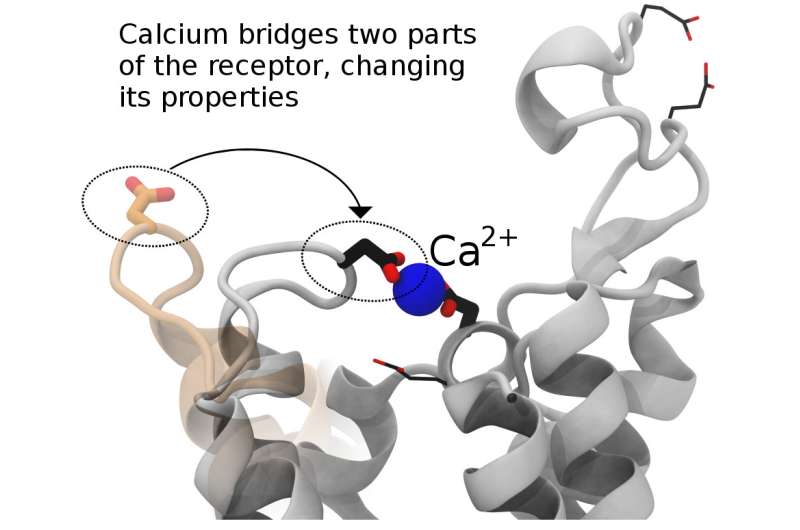
"It's a huge field of active research in academia and industry because if we can figure out precisely how GPCRs work, then we can more easily design drugs to change their behavior and thereby control pain, hunger, and more," said coauthor Christopher Neale, a researcher with the Center for Nonlinear Studies at Los Alamos National Laboratory. "This work helps us in understanding the receptors' function as a means to enable future drug discovery. For instance, if calcium binding can turn off a GPCR, then one may use that knowledge in a guided search for drugs that either promote or inhibit calcium binding depending on the desired health outcome."
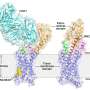
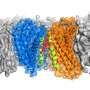
GPCRs are a family of membrane proteins that transmit information into our cells. They respond to things like adrenaline and opioid drugs, and they are the largest class of human drug targets. The research reported this week in Nature Communications describes the regulation of GPCRs by physiological ions such as sodium, calcium and magnesium.
The paper outlines in vitro nuclear magnetic resonance experiments done by Scott Prosser's group (University of Toronto, Canada) that identified changes in these receptors based on divalent cation concentrations. The paper includes confirmation of these effects in live cells by Roger Sunahara's group (University of California, San Diego) and computer simulations run at Los Alamos by Neale and Angel E. Garcia to define atomic-resolution mechanisms that can explain the experimental results. Finally, additional theory by Adnan Sljoka (Kwansei Gakuin University, Japan) showed that the mechanisms that Neale proposed involve feasible types of structural communication through the receptor.
More information: Libin Ye et al, Mechanistic insights into allosteric regulation of the A2A adenosine G protein-coupled receptor by physiological cations, Nature Communications (2018). DOI: 10.1038/s41467-018-03314-9
Journal information: Nature Communications
Provided by Los Alamos National Laboratory