Grid-based continual analysis of molecular interior for drug discovery, QSAR and QSPR
A series of grid-based computational technologies for in silico virtual screening and molecular design of new drugs is proposed. The technologies are based on original CoMIn (Continual Molecular Interior analysis) software.
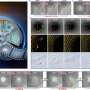
The grid-based analysis is done by means of a lattice construction, analogous to many other grid-based methods. Further continual elucidation of molecular structure is performed in various ways:
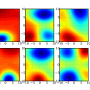
- In terms of intermolecular interactions potentials. This can be represented as a superposition of Coulomb, Van der Waals interactions and hydrogen bonds. All the potentials are well known continual functions and their values can be determined in all lattice points for a molecule.
- In terms of quantum functions such as electron density distribution, Laplacian and Hamiltonian electron density distribution, potential energy distribution, the highest occupied and the lowest unoccupied molecular orbitals distribution and their superposition.
To reduce the calculation time using quantum methods based on first principles, an original quantum free-orbital approach AlteQ is proposed. All the functions can be calculated using a quantum approach at a sufficient level of theory and their values can be determined in all lattice points for a molecule.
Then, the molecules of a dataset can be superimposed in the lattice for the maximal coincidence (or minimal deviations) of the potentials (i) or the quantum functions (ii). The methods and criteria of the superimposition are discussed. Then, a functional relationship between biological activity or property and characteristics of potentials (i) or functions (ii) is created. The methods of the quantitative relationship construction are discussed. New approaches for rational virtual drug design based on intermolecular potentials and quantum functions are invented.
More information: Andrey V. Potemkin et al, Grid-based Continual Analysis of Molecular Interior for Drug Discovery, QSAR and QSPR, Current Drug Discovery Technologies (2017). DOI: 10.2174/1570163814666170207144018
All the invented methods are explained on the http://www.chemosophia.com website.
Provided by Bentham Science Publishers