Detailed study reveals genes are constantly rearranged by cells
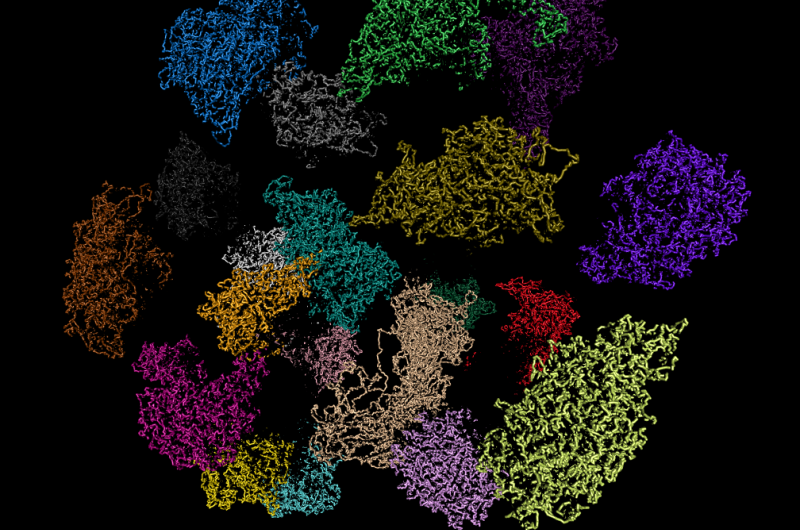
Moving genes about could help cells to respond to change according to scientists at the Babraham Institute in Cambridge, UK and the Weizmann Institute, Israel. Changing the location of a gene within a cell alters its activity. Like mixing music, different locations can make a gene 'louder' or 'quieter', with louder genes contributing more actively to the life of a cell.
Contrary to expectations, this latest study reveals that each gene doesn't have an ideal location in the cell nucleus. Instead, genes are always on the move. Published in the journal Nature, researchers examined the organisation of genes in stem cells from mice. They revealed that these cells continually remix their genes, changing their positions as they progress though different stages. This work, which has also inspired a musical collaboration, suggests that moving genes about in this way could help cells to fine-tune the volume of each gene to suit the cell's needs.
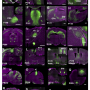
Scientists had believed that the location of genes in cells are relatively fixed with each gene having it's rightful place. Different types of cells could organise their genes in different ways, but genes weren't thought to move around much except when cells divide. This is the first time that gene organisation in individual cells has been studied in detail. The results provide snapshots of gene organisation, with each cell arranging genes in unique ways.
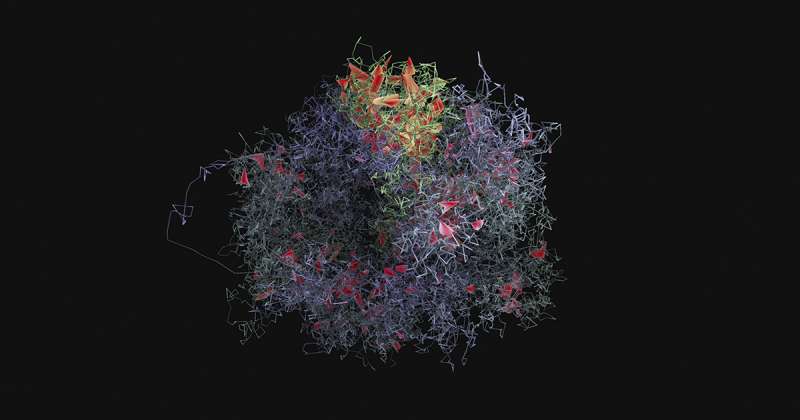
Co-first authors, Dr Takashi Nagano in the UK and Yaniv Lubling in Israel have collected and individually analysed information one-by-one from over 4000 cells for this study. Speaking about the work, Dr Nagano said: "We've never had access to this level of information about how genes are organised before. Being able to compare between thousands of individual cells is an extremely powerful tool and adds an important dimension to our understanding of how cells position their genes."
Collecting hundreds of thousands of pieces of information about gene positions from just one cell is a significant challenge and it has relied on technology pioneered by Dr Nagano and colleagues in 2013. Combining this technology, called single-cell Hi-C, with statistical analyses performed by Dr Amos Tanay's team at the Weizmann Institute has made this detailed research possible. A version of the Hi-C technique was also recently shown to have the potential to improve cancer diagnoses.
Lead author from the Babraham Institute, Professor Peter Fraser said: "We typically see that changes to gene activity have a great impact on health, disease and evolution. It's now obvious that genome organisation may have a part to play in this and our research shows that the effects of location on genes may be a constantly moving target. Understanding how the genome is controlled during this constant re-shuffling is an important step towards understanding how our genomes and genes effect our lives."
The team now plan to examine whether changing the locations of genes actually has a significant effect on the volume of each gene and to study different types of cells to understand whether they move genes about less once they stop dividing or if all cells behave like stem cells do.
This research has previously inspired an artistic collaboration with music producer Max Cooper and visual artist Andy Lomas, who produced the music tracks 'Chromos' and 'Coils of Living Synthesis'. The accompanying video was based on computational models of DNA folding generated by author Dr Csilla Varnai.
More information: Takashi Nagano et al, Cell-cycle dynamics of chromosomal organization at single-cell resolution, Nature (2017). DOI: 10.1038/nature23001
Journal information: Nature
Provided by Babraham Institute