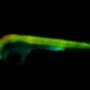
Shedding light on protein interaction networks in a developing organism

Researchers succeeded for the first time in mapping protein-protein interactions in living developing plant roots. The findings of three international research groups led by the department of Plant Developmental Biology at Wageningen University and Research were published in Nature.
The work is considered a breakthrough in field of cell and developmental biology as interactions between proteins at physiological conditions are for the first time visualized within intact living tissue in a non-invasive manner. The results using this approach will allow a better understanding of protein-binding events which are essential for many important cellular functions as their dis-function is causal of diseases including cancer.
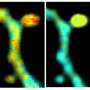
A joint collaboration of Wageningen University & Research, Amsterdam University and Düsseldorf University succeeded in mapping protein interactions in living plant roots at native conditions and at cellular resolution using the latest state-of-the-art functional imaging technologies. The recently published work in Nature titled "In vivo FRET-FLIM reveals cell type-specific protein interactions in Arabidopsis roots" combined FRET-FLIM (Förster Resonance Energy Transfer) –Fluorescence Lifetime imaging Microscopy) technology with genetics and gene expression analysis. The work unraveled that in living tissues, protein complexes can change their conformation in a cell type dependent manner to regulate specific gene expression programs leading to precise specification and maintenance of particular cell fates within the Arabidopsis root meristem.
Understanding root growth
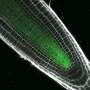
At the tip of a growing plant root resides a population of "stem cells" These cells divide and differentiate into different cell types generating different tissues in the plant root. The fate of each cell, hence what kind of root tissue it will generate, is determined by combinatory modules where, transcription factors play a crucial role. To understand how these proteins regulate cell fate and consequently root growth Ikram Blilou, together with her PhD student Yuchen Long, faced the challenge to visualize the interactions between a set of key transcription factors focusing on how these interactions change in time and space in different tissue types and how they regulate cell fate during root development
Technical challenge
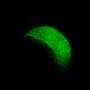
It is technically challenging to study protein interactions in living organisms because of their low abundance, making it difficult detect these interactions by traditional biochemical methods. In this study the researchers used state-of-the-art functional imaging and molecular biology techniques to visualize specific interactions using FRET-FLIM (technique in combination with genetics and gene expression analysis. Proteins interact by direct binding, this means they come very close to each other and in in this case FRET techniques is used to detect these interactions by tagging the proteins of interest with genetically encoded fluorescent proteins.
Combined expertise
This study is the result of a successful collaboration between three international teams, in addition to Plant Developmental Biology group at Wageningen University headed by Prof Ben Scheres, the work involved Prof. Theodorus Gadella from the Van Leeuwenhoek Centre for Advanced Microscopy in Amsterdam. The team of Gadella, including Joachim Goedhart and Marten Postma, has a long standing expertise in optimization of fluorescent proteins, quantitative microscopy and advanced FRET-FLIM analysis. Together with the team of Rüdiger Simon from Düsseldorf University, including Yvonne Stahl and Stephanie Stefanie-Weidtkamp-Peters with expertise on high resolution life time imaging, the FRET-FLIM technology was implemented overcoming the current limitations for studying protein complex dynamics at cellular resolution.
More information: Yuchen Long et al. In vivo FRET–FLIM reveals cell-type-specific protein interactions in Arabidopsis roots, Nature (2017). DOI: 10.1038/nature23317
Journal information: Nature
Provided by Wageningen University