Thousands of images of frozen bacteria

How do bacteria sense and adapt to their environment? Ariane Briegel, Professor of Ultrastructural Biology, is intrigued by this question. Using new techniques, she produces three-dimensional images of bacteria that provide us with new clues about their sensory system.
Have you always been interested in bacteria?
'Actually, no. When I was a student, what I really wanted to do was marine biology. But I lived in Munich, which is about as far away from any ocean you can get. So I decided to join the zoology department instead. I did a lot of microscopy there, and became fascinated by it: every time you look through the microscope, you see something completely new. After completing my master's, I started a PhD at the Max Planck Institute, where I learned about the new microscopy technique of electron cryotomography. I've been working on bacteria ever since, and I never regreted it. It's such an exciting field, there's so much to discover.'
What is your main research interest?
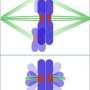
'What fascinates me most, is how bacteria detect signals from their environment. How can they find sugars, avoid toxins and adapt to changing environments? We already know that bacteria use certain signals, like the presence of particular chemical substances, to determine where they need to go. But how do they detect those signals? What does their 'nose' look like? The images we make with electron cryotomography show us that many types of bacteria have large chemo receptors. These help them detect sugars in the environment, for instance.'
Could you explain what electron cryotomography is?
'It's a relatively new technique, which allows us, for the first time, to look at bacteria in their natural state, at molecular resolution and in three dimensions. Electron microscopy (EM) relies on electrons to form an image. Since the wavelength of electrons is much shorter than that of light, this method can achieve much higher resolutions than light microscopy. But in order to use electrons to generate an image, you have to work in a vacuum so the electrons dont get scattered by the atmosphere. Biological material is full of water which has to be removed before such a sample can be imaged with traditional EM, since the water of the sample would evaporate in the vacuum and destroy the sample.
Instead of removing the water and changing the structure of the samples, we flash freeze the cells. The freezing has to happen so fast that water cannot form ice crystals. This keeps the cells intact and allows imaging of nearly artifact-free samples. Additionally, we collect a large set of images for each sample by rotating it in the microscope. This produces many views of the cell from different angles. From those, we computationally generate a 3-D-view of the cells.'
What are the applications of this type of research?
'The insights we gain in bacteria may provide a foundation for the development of new treatments against diseases. If we find out how potentially harmful bacteria use their ability to sense and respond to their environment in order to invade a host, that knowledge may help us prevent infections. We are now starting a research project on v.cholerae, the microbe that causes cholera, which may provide helpful insights for the development of new cholera treatments. And there are other possible applications as well, such as using bacteria to get rid of toxic waste. But my research is essentially fundamental: I want to know exactly how the sensory system of bacteria works. Who knows what it will be used for in the future.'
Do you often make use of the equipment of the Netherlands Center for Electron Nanoscopy (NeCEN)?
'Yes, I absolutely rely on it. In fact, NeCEN is the main reason I came to Leiden. It's a great facility for electron cryotomography, since it has two of the best microscopes that are currently on the market. It also has a great support staff that provides expert support on site. We grow cells and prepare the samples in our own lab, but all of the imaging happens at NeCEN.'
This type of imaging generates a whole lot of data, I imagine?
'Yes, definitely. Each dataset consists of 121 high resolution images, which can be split into 10 subframes each. The data accumulates very quickly; in my previous lab, we collected about 30.000 of those datasets during my time there. Here in Leiden we are just getting started with data collection, so we are now in the process of setting up a data management system. Dealing with all this data in is going to be a big issue for us, so I hope to collaborate with the Computer Science department for that.'
How do you analyse all this data?
'Right now, we process each dataset by hand. But we are planning on using software that can do most of it automatically: processing the data, reconstructing it and generating videos. In my previous lab, we had a nice system set up for that, which saved us a great deal of time.'
How do you think your field will develop in the future?
'One of the biggest limitations in electron crytomography so far is that the samples you use have to be really thin, since the electron beam has to be able to pass through it. That is why a lot of work has been done on bacteria, which are naturally thin and small. But the development of new techniques will allow us to freeze larger samples, such as human cells and tissues, and then slice then samples very thinly to allow imaging with EM. This will open up a whole new field with many possibilities for research. This summer we'll get such a machine at the IBL, which will allow us to image thicker samples than individual bacteria. Then we will be able to look at host-microbe interactions, so I am very excited about that.'
Provided by Leiden University