New structural studies reveal workings of a molecular pump that ejects cancer drugs
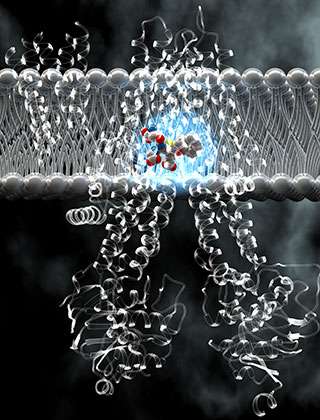
Sometimes cells resist medication by spitting it back out. Cancer cells, in particular, have a reputation for defiantly expelling the chemotherapy drugs meant to kill them. Researchers at The Rockefeller University have shed new light on a molecular pump that makes this possible, by determining its three-dimensional structure, down to the level of atoms.
"This molecular machine ejects numerous anticancer agents, as well as other drugs. However, no one understood how it can recognize and remove such an impressive variety of substances," says lead researcher Jue Chen, the William E. Ford Professor and head of the Laboratory of Membrane Biology and Biophysics.
"By examining how this drug resistant pump binds to its cargo before transporting it, we have found an answer," she adds. The new structures, described February 23 in Cell, could help to guide the development of more effective treatments for cancer and other disorders.
Mysterious versatility
Known as MRP1, the pump in question was identified in drug-resistant lung cancer cells in 1992. While some cancer cells express an unusual abundance of this protein, it is also common within normal cells. It's a part of the barrier that protects the brain from infection, and helps export hormones, immune signaling compounds, and other cargo, including unwanted foreign substances. Unfortunately for modern medicine, MRP1 often mistakes useful chemicals, including opiates, antidepressants, and antibiotics, for potentially harmful ones in need of removal.
Proteins charged with transportation across a cell's membrane tend to be picky, accepting only particular types of cargo, in some cases a single molecule. MRP1 is different, but because researchers couldn't get a detailed look at it, they couldn't be sure why. That's because the standard method for determining the structure of a molecule required them to turn it into crystals, something they couldn't do with MRP1.
Chen and study researcher Zachary Johnson, a postdoc in her lab who performed this research, got around this problem using an imaging approach known as electron cryomicroscopy, in which molecules are frozen in solution rather than crystallized. With help from Mark Ebrahim and Johanna Sotiris in Rockefeller's Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center, they captured images of MRP1 alone and bound to its cargo.
A two-part pocket
Deep inside MRP1, they found the key to its versatility: the pocket in which it clasps its cargo. Formed by the two sides of the pump, this pocket's structure is flexible, allowing it to accommodate cargo of varying size. It also has two halves, each with distinct chemical properties. As it turns out, many of the substances that bind to MRP1 match the pocket's two-part footprint; those that don't are coupled to a filler molecule from inside the cell.
Once in the pocket, the cargo connects the two halves of MRP1, pulling them together, as if pinching the prongs of a clothespin, Chen and Johnson saw. This movement serves as the first step in changing the shape of the pump so that the cargo can be released outside the cell.
A template for improvement
MRP1 is part of a large family of pumps involved in many physiological processes throughout the body. Since other members of this family also eject drugs, this work has medical relevance beyond MRP1.
"Ultimately, we hope these and other structures like them could serve as templates for designing chemotherapy drugs the pumps don't recognize, or inhibitors that block them," Johnson says. "It's possible that such a strategy could also improve the effectiveness of other drugs, such as those that need to reach the brain."
More information: Zachary Lee Johnson et al, Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1, Cell (2017). DOI: 10.1016/j.cell.2017.01.041
Journal information: Cell
Provided by Rockefeller University