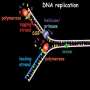
Molecular hairpin structures make effective DNA replicators

The evolution of cells and organisms is thought to have been preceded by a phase in which informational molecules like DNA could be replicated selectively. New work shows that hairpin structures make particularly effective DNA replicators.
In the metabolism of all living organisms there is a clear division of labor: Nucleic acids (DNA and RNA) carry the information for the synthesis of proteins, and proteins provide the structural and executive functions required by cells, such as the controlled and specific catalysis of chemical reactions by enzymes. However, in recent decades, it has become clear that this distinction is by no means absolute. In particular RNA is capable of ignoring the boundary outlined above and is known to play a catalytic role in many important processes. For example, certain RNA molecules can catalyze the replication of other nucleic acids, and this versatility could help to explain how life originated on Earth.
Nucleic acid molecules are made up of subunits called nucleotides, which differ in their so-called bases. The bases found in RNA are referred to as A, C, G and U (DNA uses T in place of U). These bases fall into two complementary pairs, whose members specifically interact, A with T (or U) and G with C. This complementarity is what accounts for the stability of the DNA double helix, and enables single strands of RNA to fold into complex shapes.
Life is thought to have emerged from a process of chemical evolution in which nucleic acid sequences could be selectively replicated. Thus, in prebiotic systems certain molecular "species" that carried information were reproduced at the expense of others. In biological systems, such selectivity is normally mediated by so-called primers—strands of nucleic acid that pair (as described above) with part of the molecule to be replicated, to form a short double helix. The primer provides a starting point for the extension of the double-stranded region to form a new daughter strand. Moreover, this process can be reconstructed in the test-tube.
The pros and cons of hairpin replicators
Georg Urtel and Thomas Rind, who are members of the research group led by Dieter Braun (Professor of Systems Biophysics at LMU), have used such a system to identify properties the might favor the selective replication of DNA molecules. For their experiments, they chose a single-stranded DNA sequence that adopts a so-called hairpin structure. In these molecules, the base sequences at either end are complementary to each other, as are short stretches of sequence within the rest of the molecule. This distribution of complementary sequences causes such a strand to fold into a hairpin-like conformation.
Thanks to the pairing rules outlined above, replication of a single strand of DNA produces a second strand whose sequence differs from that of the first. Each strand of a non-hairpin structure therefore needs its own primer for replication. But with hairpins, one primer suffices to prime synthesis of both the original and its complementary strand. "This means that hairpins are relatively simple replicators," Georg Urtel points out. The downside is that the hairpin structure makes primer binding more difficult, and this in turn limits their replication rate. Molecular species that are devoid of hairpin structures don't have this problem.
Cooperation beats competition
In subsequent experiments the researchers discovered that two simple hairpin species could cooperate to give rise to a much more efficient replicator, which requires two primers for its amplification. The two hairpin species selected each required a different primer, but their sequences were in part identical. The switch to cooperative replication occurs when replication of one of the hairpins stalls. "As a rule, replication processes in nature are never perfect," says Dieter Braun. "Such a premature halt is not something that one needs to design into the system. It happens stochastically and we make use of it in our experiments." The partially replicated hairpin can, however, bind to a molecule of the second species, and serves as a primer that can be further elongated. Moreover, the resulting product no longer forms a hairpin. In other words, it represents a new molecular species.
Saved from extinction
Such so-called 'crossbreeds' need two primers for their replication, but can nevertheless be replicated significantly faster than either of their hairpin progenitors For further experiments showed that, upon serial dilution of the population, the hairpin DNAs soon become extinct. However, the sequence information they contained survives in the crossbreeds and can be replicated further.
The converse experiment confirmed that information is indeed conserved: If crossbreeds are supplied with only one primer, the corresponding progenitor hairpin species can still be replicated by the kind of switching process mentioned above. But, in the absence of the second primer, the crossbreed dies out. "Thus, the crossbreeding process not only provides for the transition from 'simple and slow' replicators to more rapid replicators, it also makes it possible for the system to adapt to the prevailing conditions," Urtel explains. "It also suggests how early replicators could have cooperated with each other under prebiotic conditions prior to the origin of living systems."
More information: Georg C. Urtel et al. Reversible Switching of Cooperating Replicators, Physical Review Letters (2017). DOI: 10.1103/PhysRevLett.118.078102
Journal information: Physical Review Letters
Provided by Ludwig Maximilian University of Munich