A comprehensive comparison of methodologies that quantify RNAs of single cells
Every cell has its own individual molecular fingerprint, which is informative for its functions and regulatory states. LMU researchers have now carried out a comprehensive comparison of methodologies that quantify RNAs of single cells.
The cell is the fundamental unit of all living organisms. Hence, in order to understand essential biological processes and the perturbations that give rise to disease, one must first dissect the functions of cells and the mechanisms that regulate them. Modern high-throughput protein and nucleic-acid sequencing techniques have become an indispensable component of this endeavor. In particular, single-cell RNA sequencing (scRNA-seq) permits one to determine the levels of RNA molecules – the gene copies - that are expressed in a given cell, and several versions of the methodology have been described in recent years. The spectrum of genes expressed in a given cell amounts to a molecular fingerprint, which yields a detailed picture of its current functional state. "For this reason, the technology has become an extraordinarily valuable tool, not only for basic research but also for the development of new approaches to treat diseases," says LMU biologist Wolfgang Enard. Enard and his team have now undertaken the first comprehensive comparative analysis of the various RNA sequencing techniques, with regard to their sensitivity, precision and cost efficiency. Their results appear in the leading journal Molecular Cell.
The purpose of scRNA-seq is to identify the relative amounts of the messenger RNA (mRNA) molecules present in the cells of interest. mRNAs are the blueprints that specify the structures of all the proteins made in the cell, and represent "transcribed" copies of the corresponding genetic information encoded in specific segments of the genomic DNA in the cell nucleus. In the cytoplasm surrounding the nucleus, the nucleotide sequences of mRNAs are "translated" into the amino-acid sequences of proteins by molecular machines called ribosomes. Thus a complete catalog of the mRNAs in a cell provides a comprehensive view of the proteins that it produces, and tells one what subset of the thousands of genes in the genome are active and how their activity is regulated. Furthermore, aberrant patterns of gene activity point to disturbances in gene expression and cell function, and reveal the presence of specific pathologies. The scRNA-seq procedure itself can be carried out using commercially available kits, but many researchers prefer to assemble the components required for their preferred formulations themselves.
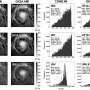
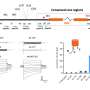
In order to ascertain which of the methods currently in use is most effective and economical, Enard and his colleagues applied six different methods to mouse embryonal stem cells and compared the spectra of mRNAs detected by each of them. They then used this data to compute how much it costs for each method to reliably detect differently expressed genes between two cell types. "This comparison revealed that some of the commercial kits are ten times more expensive than the corresponding home-made versions," Enard says. However, the researchers point out that the choice of the optimal method largely depends on the conditions and demands of the individual experiment. "It does make a difference whether one wants to analyze the activity of hundreds of genes in thousands of individual cells, or thousands of genes in hundreds of cells," Enard says. "We were able to demonstrate which method is best for a given purpose, and we also obtained data that will be useful for the further development of the technology."
The new findings are of particular interest in the field of genomics. For example, scRNA-seq is a fundamental prerequisite for the success of the effort to assemble a Human Cell Atlas – one of the most ambitious international projects in genomics since the initial sequencing of the human genome. It aims to provide no less than a complete inventory of all the cell types and subtypes in the human body at all stages of development from embryo to adult on the basis of their patterns of gene activity. It is estimated that the total number of cells in the human body is on the order of 3.5 × 1013. Scientists expect that such an atlas would revolutionize our knowledge of human biology and our understanding of disease processes.
More information: Christoph Ziegenhain et al. Comparative Analysis of Single-Cell RNA Sequencing Methods, Molecular Cell (2017). DOI: 10.1016/j.molcel.2017.01.023
Journal information: Molecular Cell
Provided by Ludwig Maximilian University of Munich