Slo-mo unwrapping of nucleosomal DNA probes protein's role
Nucleosomes are tightly packed bunches of DNA and protein which, when linked together as chromatin, form each of the 46 chromosomes found in human cells.
The organization of DNA in nucleosomes is important not just for DNA packaging; it also forms the basis for the regulation of gene expression. By controlling the access to DNA, nucleosomes help facilitate all kinds of gene activity, from RNA transcription to DNA replication and repair.
A research group led by Lois Pollack, professor of applied and engineering physics, used a combination of X-ray and fluorescence-based approaches to study how the shapes and compositions of nucleosomes change after being destabilized.
The group's paper, "Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core," is published online in Proceedings of the National Academy of Sciences. Co-lead authors are postdoctoral researcher Yujie Chen and doctoral student Joshua Tokuda.
Among the collaborators was Lisa Gloss of the Washington State University School of Molecular Biosciences. Gloss' group conducted NCP analysis using Förster resonance energy transfer (FRET) measurements.
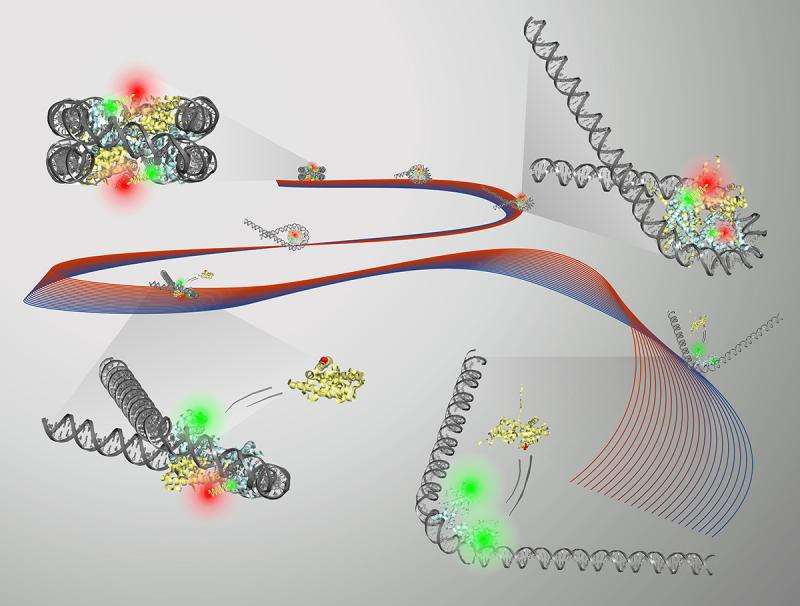
The nucleosome core particle (NCP) consists of DNA wrapped around a core of eight histone proteins. Access to DNA is regulated through the unwrapping of the histone core, and an understanding of how this remodeling occurs can inform therapeutic strategies for many diseases, including cancer.
In the body, unwrapping of the DNA structure is triggered by proteins, but Pollack and her group used a salt solution. Earlier work by the group, published in 2014, developed a new way to show that a high-concentration salt solution could make the DNA inside the nucleosome unwrap.
This time around, to slow the unwrapping process down, the group used a salt solution that was about one-third less concentrated. "We discovered that by using a lower salt concentration, we were able to slow down the disassembly process and gain new insights into how the DNA unwraps," Tokuda said.
Using FRET, small-angle X-ray scattering and other methods, the group was able to get a clear picture of the DNA activity during unwrapping of the histone core. It was found that different DNA shapes were produced during the unwrapping process, most notably a "teardrop" shape that seemed to promote protein activity.
The histone core goes from eight protein molecules to six when the DNA unwraps into the teardrop shape. "It's as if having the DNA in this shape is a signal to the protein: 'Hey, now's the time. You want to change it up? Go ahead,'" Pollack said.
This finding suggests that the molecular transition is guided by this specific type of unwrapping. It's a step toward better understanding of DNA access during transcription, replication and repair.
"The reason why these structures are so important, in addition to packaging, is that it also gives cells the opportunity to control which genes are on and off," Tokuda said.
Tokuda adds that misregulation of chromatin remodeling is also implicated in many human diseases, from neuro-development and degenerative disorders to immunodeficiency syndromes and cancer.
"We hope that by developing these tools to investigate the fundamental mechanism of remodeler proteins," he said, "we may be able to provide insight that will aid in the development of new therapeutic strategies for these diseases."
More information: Yujie Chen et al. Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1611118114
Journal information: Proceedings of the National Academy of Sciences
Provided by Cornell University