Expanded genetic information about the migratory locust

Researchers from the University of Granada (UGR) have expanded the knowledge of genetic information about the migratory locust (Locusta migratoria), an orthopteron responsible for devastating plagues in Africa due to its high mobility, capable of flying at high speed with the wind (up to 20 kilometers per hour).
The scientists of the Department of Genetics have discovered for the first time 62 satellite DNA (satDNA) families in this species thanks to the development of a computer protocol (satMiner) based on the bioinformatic programs Repeat Explorer and DeconSeq. The authors have also found 85 satellite DNA families in the Luzula elegans plant, comprising more than twice the families previously described by a conventional analysis, and with abundances over 20 times lower.
The UGR researchers Francisco J. Ruiz-Ruano Campaña, Maria Dolores López León, Josefa Cabrero and Juan Pedro Martínez Camacho applied satMiner to the search for satellites in the migratory locust, a species with no previously known satellite DNA.
In a paper published in the journal Scientific Reports, scientists from the UGR have also proposed a new scientific term, "satellitome," to refer to the complete catalog of satellite DNAs that exist in a given genome, and they have developed methods to study it in depth.
Until recently, it was thought that eukaryotic genomes contained only a few types of satellite DNA in the heterochromatin of chromosomes—that is, in the most condensed regions. There had only been a few cases in which more than two or three satellites were described in the same species.
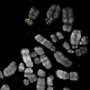
The physical mapping of satDNAs in the genome of the migratory locust is an important milestone, since nobody had described so many satellites in a species before. This has allowed them to discover that the distribution of satDNAs on the chromosomes can be dispersed or clustered—that is to say, highly concentrated in some chromosomal regions—independent of the length of the repeating monomer. And more than half of the satellites are clustered on a single chromosome; surprisingly, they were present in tens or hundreds of contigs (sets of overlapping DNA segments that represent a consensus region of DNA). These contigs were different from the locust genome, which suggests that they are scattered around the genome.
The authors of this research expect that the in-depth analysis of satellitomes in other species by these new methodologies opens new lines of research about the evolution of satellite DNAs in intra- and interspecific levels, and that the use of satDNAs as markers of chromosome identification may be helpful to complete genomic sequencing currently in progress.
Finally, the UGR scientists hope the discovery of so many new satellites helps find new features for this largely unknown part of the chromosome.
More information: Francisco J. Ruiz-Ruano et al, High-throughput analysis of the satellitome illuminates satellite DNA evolution, Scientific Reports (2016). DOI: 10.1038/srep28333
Journal information: Scientific Reports
Provided by University of Granada